Metabolites modulate the functional state of human uridine phosphorylase I
- PMID: 32864839
- PMCID: PMC7586905
- DOI: 10.1002/pro.3939
Metabolites modulate the functional state of human uridine phosphorylase I
Abstract
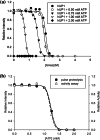
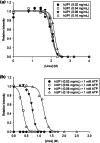
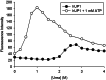
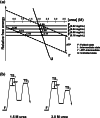
Metabolic pathways in cancer cells typically become reprogrammed to support unconstrained proliferation. These abnormal metabolic states are often accompanied by accumulation of high concentrations of ATP in the cytosol, a phenomenon known as the Warburg Effect. However, how high concentrations of ATP relate to the functional state of proteins is poorly understood. Here, we comprehensively studied the influence of ATP levels on the functional state of the human enzyme, uridine phosphorylase I (hUP1), which is responsible for activating the chemotherapeutic pro-drug, 5-fluorouracil. We found that elevated levels of ATP decrease the stability of hUP1, leading to the loss of its proper folding and function. We further showed that the concentration of hUP1 exerts a critical influence on this ATP-induced destabilizing effect. In addition, we found that ATP interacts with hUP1 through a partially unfolded state and accelerates the rate of hUP1 unfolding. Interestingly, some structurally similar metabolites showed similar destabilization effects on hUP1. Our findings suggest that metabolites can alter the folding and function of a human protein, hUP1, through protein destabilization. This phenomenon may be relevant in studying the functions of proteins that exist in the specific metabolic environment of a cancer cell.
Keywords: ATP; ligand-binding; protein functional state; protein stability; uridine phosphorylase I.
© 2020 The Protein Society.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

