Human NK cell deficiency as a result of biallelic mutations in MCM10
- PMID: 32865517
- PMCID: PMC7524476
- DOI: 10.1172/JCI134966
Human NK cell deficiency as a result of biallelic mutations in MCM10
Abstract
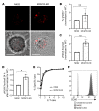
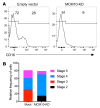
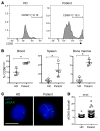
Human natural killer cell deficiency (NKD) arises from inborn errors of immunity that lead to impaired NK cell development, function, or both. Through the understanding of the biological perturbations in individuals with NKD, requirements for the generation of terminally mature functional innate effector cells can be elucidated. Here, we report a cause of NKD resulting from compound heterozygous mutations in minichromosomal maintenance complex member 10 (MCM10) that impaired NK cell maturation in a child with fatal susceptibility to CMV. MCM10 has not been previously associated with monogenic disease and plays a critical role in the activation and function of the eukaryotic DNA replisome. Through evaluation of patient primary fibroblasts, modeling patient mutations in fibroblast cell lines, and MCM10 knockdown in human NK cell lines, we have shown that loss of MCM10 function leads to impaired cell cycle progression and induction of DNA damage-response pathways. By modeling MCM10 deficiency in primary NK cell precursors, including patient-derived induced pluripotent stem cells, we further demonstrated that MCM10 is required for NK cell terminal maturation and acquisition of immunological system function. Together, these data define MCM10 as an NKD gene and provide biological insight into the requirement for the DNA replisome in human NK cell maturation and function.
Keywords: Genetic diseases; Immunology; NK cells.
Conflict of interest statement
Figures
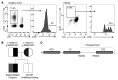
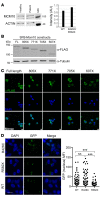
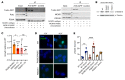

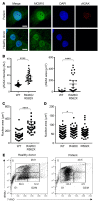

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

