Cloning, isolation, and characterization of novel chitinase-producing bacterial strain UM01 (Myxococcus fulvus)
- PMID: 32865699
- PMCID: PMC7458996
- DOI: 10.1186/s43141-020-00059-1
Cloning, isolation, and characterization of novel chitinase-producing bacterial strain UM01 (Myxococcus fulvus)
Abstract
Background: Chitin is an important biopolymer next to cellulose, extracted in the present study. The exoskeleton of marine bycatch brachyuran crabs, namely Calappa lophos, Dromia dehaani, Dorippe facchino and also from stomatopod Squilla spp. were used to extract chitin through fermentation methods by employing two bacterial strains such as Pseudomonas aeruginosa, Serratia marcescens. The yield of chitin was 44.24%, 37.45%, 11.56% and 27.24% in C. lophos, D. dehaani, D. facchino and Squilla spp. respectively. FT-IR spectra of the produced chitin exhibit peaks which is more or less coherent to that of standard chitin which is further analysed by Scanning Electron Microscope. The quality of produced chitin was assessed through moisture, protein, ash and lipid content analysis ensured that chitin obtained from trash crustaceans are on par with that of standard chitin.
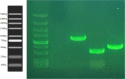
Results: A total of 10 samples were collected from different areas of Jiangsu China for screening of chitinase-producing bacteria. Based on the clearance zone, two of the best samples were chosen for further study. 16S rRNA sequence analysis showed that this strain belongs to genus Myxococcus and species Myxococcus fulvus. Phylogenetic analysis was performed and it shows strain UM01 is a novel bacterial strain. UM01 isolate shows maximum chitinase production at 35 °C and 8 pH. Among all, these colloidal chitins were found to be the best for chitinase production. Three chitinase-producing genes were identified and sequenced by using degenerative plasmid. UMCda gene (chitin disaccharide deacetylase) was cloned into E. coli DH5a by using PET-28a vector, and antagonistic activity was examined against T. reesei.
Conclusion: To our knowledge, this is the earliest study report to gene cloning and identification of the chitinase gene in Myxococcus fulvus. Chitinase plays a key role in decomposition and utilization of chitin as a raw material. This research indicates that Myxococcus fulvus UM01 strain is a novel myxobacteria strain and can produce large amounts of chitinase within a short time. The UMCda gene cloned into E. coli DH5a showed a promising effect as antifungal activity. In overall findings, the specific strain UM01 has endowed properties of bioconversation of waste chitin and other biological applications.
Keywords: Chitinase; Gene cloning; Myxococcus fulvus.
Conflict of interest statement
The authors declare that they have no competing interests
Figures









References
-
- Aideia 3 ZE 2 EM 1; N l-D, Hoda AM. Using of chitosan as antifungal agent in Kariesh cheese. New York Sci J 2012;5(9)
-
- Muzzarelli RAA, Boudrant J, Meyer D, et al. Current views on fungal chitin/chitosan, human chitinases, food preservation, glucans, pectins and inulin: a tribute to Henri Braconnot, precursor of the carbohydrate polymers science, on the chitin bicentennial. Carbohydr Polym. 2012;87:995–1012. doi: 10.1016/j.carbpol.2011.09.063. - DOI
-
- Rudall KM, Kenchington W. The chitin system. Biol Rev. 1973;48:597–633. doi: 10.1111/j.1469-185x.1973.tb01570.x. - DOI
-
- Rudall KM. Chitin and its association with other molecules. J Polym Sci Part C Polym Symposia. 2007;28:83–102. doi: 10.1002/polc.5070280110. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
