Crossing complexity of space-filling curves reveals entanglement of S-phase DNA
- PMID: 32866178
- PMCID: PMC7458320
- DOI: 10.1371/journal.pone.0238322
Crossing complexity of space-filling curves reveals entanglement of S-phase DNA
Abstract
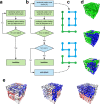
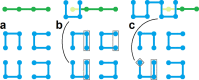
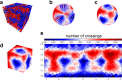
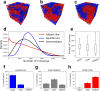
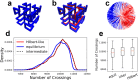
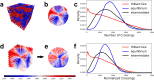
Space-filling curves have been used for decades to study the folding principles of globular proteins, compact polymers, and chromatin. Formally, space-filling curves trace a single circuit through a set of points (x,y,z); informally, they correspond to a polymer melt. Although not quite a melt, the folding principles of Human chromatin are likened to the Hilbert curve: a type of space-filling curve. Hilbert-like curves in general make biologically compelling models of chromatin; in particular, they lack knots which facilitates chromatin folding, unfolding, and easy access to genes. Knot complexity has been intensely studied with the aid of Alexander polynomials; however, the approach does not generalize well to cases of more than one chromosome. Crossing complexity is an understudied alternative better suited for quantifying entanglement between chromosomes. Do Hilbert-like configurations limit crossing complexity between chromosomes? How does crossing complexity for Hilbert-like configurations compare to equilibrium configurations? To address these questions, we extend the Mansfield algorithm to enable sampling of Hilbert-like space filling curves on a simple cubic lattice. We use the extended algorithm to generate equilibrium, intermediate, and Hilbert-like configurational ensembles and compute crossing complexity between curves (chromosomes) in each configurational snapshot. Our main results are twofold: (a) Hilbert-like configurations limit entanglement between chromosomes and (b) Hilbert-like configurations do not limit entanglement in a model of S-phase DNA. Our second result is particularly surprising yet easily rationalized with a geometric argument. We explore ergodicity of the extended algorithm and discuss our results in the context of more sophisticated models of chromatin.
Conflict of interest statement
No, the authors have declared that no competing interests exist.
Figures




Similar articles
-
Lossless compression of medical images using Hilbert space-filling curves.Comput Med Imaging Graph. 2008 Apr;32(3):174-82. doi: 10.1016/j.compmedimag.2007.11.002. Epub 2008 Jan 8. Comput Med Imaging Graph. 2008. PMID: 18248789
-
Chromatin folding--from biology to polymer models and back.J Cell Sci. 2011 Mar 15;124(Pt 6):839-45. doi: 10.1242/jcs.077628. J Cell Sci. 2011. PMID: 21378305
-
On computational algorithms for real-valued continuous functions of several variables.Neural Netw. 2014 Nov;59:16-22. doi: 10.1016/j.neunet.2014.05.015. Epub 2014 Jun 26. Neural Netw. 2014. PMID: 25036646
-
Computational approaches from polymer physics to investigate chromatin folding.Curr Opin Cell Biol. 2020 Jun;64:10-17. doi: 10.1016/j.ceb.2020.01.002. Epub 2020 Feb 8. Curr Opin Cell Biol. 2020. PMID: 32045823 Review.
-
Models of chromosome structure.Curr Opin Cell Biol. 2014 Jun;28:90-5. doi: 10.1016/j.ceb.2014.04.004. Epub 2014 May 4. Curr Opin Cell Biol. 2014. PMID: 24804566 Review.
References
-
- Schram RD, Schiessel H. Exact enumeration of Hamiltonian walks on the 4× 4× 4 cube and applications to protein folding. Journal of Physics A: Mathematical and Theoretical. 2013;46(48):485001.
-
- Smrek J, Grosberg AY. On enumeration of Hilbert-like curves. Journal of Physics A: Mathematical and Theoretical. 2015;48(19):195001.
-
- Ramakrishnan R, Pekny J, Caruthers J. A combinatorial algorithm for effective generation of long maximally compact lattice chains. The Journal of chemical physics. 1995;103(17):7592–604.
-
- Lua R, Borovinskiy AL, Grosberg AY. Fractal and statistical properties of large compact polymers: a computational study. Polymer. 2004;45(2):717–31.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

