Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19
- PMID: 32866534
- PMCID: PMC7455547
- DOI: 10.1016/j.virusres.2020.198146
Scaffold morphing of arbidol (umifenovir) in search of multi-targeting therapy halting the interaction of SARS-CoV-2 with ACE2 and other proteases involved in COVID-19
Abstract
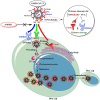
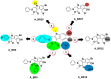
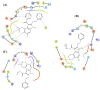
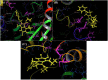
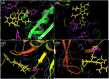
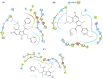
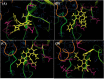
The rapid emergence of novel coronavirus, SARS-coronavirus 2 (SARS-CoV-2), originated from Wuhan, China, imposed a global health emergency. Angiotensin-converting enzyme 2 (ACE2) receptor serves as an entry point for this deadly virus while the proteases like furin, transmembrane protease serine 2 (TMPRSS2) and 3 chymotrypsin-like protease (3CLpro) are involved in the further processing and replication of SARS-CoV-2. The interaction of SP with ACE2 and these proteases results in the SARS-CoV-2 invasion and fast epidemic spread. The small molecular inhibitors are reported to limit the interaction of SP with ACE2 and other proteases. Arbidol, a membrane fusion inhibitor approved for influenza virus is currently undergoing clinical trials against COVID-19. In this context, we report some analogues of arbidol designed by scaffold morphing and structure-based designing approaches with a superior therapeutic profile. The representative compounds A_BR4, A_BR9, A_BR18, A_BR22 and A_BR28 restricted the interaction of SARS-CoV-2 SP with ACE2 and host proteases furin and TMPRSS2. For 3CLPro, Compounds A_BR5, A_BR6, A_BR9 and A_BR18 exhibited high binding affinity, docking score and key residue interactions. Overall, A_BR18 and A_BR28 demonstrated multi-targeting potential against all the targets. Among these top-scoring molecules A_BR9, A_BR18, A_BR22 and A_BR28 were predicted to confer favorable ADME properties.
Keywords: ADME; Arbidol; COVID-19; Molecular docking; SARS-CoV-2; Scaffold morphing.
Copyright © 2020 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures







References
-
- Bestle D., Heindl M.R., Limburg H., Pilgram O., Moulton H., Stein D.A., Hardes K., Eickmann M., Dolnik O., Rohde C. 2020. TMPRSS2 And Furin are Both Essential for Proteolytic Activation and Spread of SARS-CoV-2 in Human Airway Epithelial Cells and Provide Promising Drug Targets. bioRxiv. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

