Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures
- PMID: 32867141
- PMCID: PMC7563858
- DOI: 10.3390/biom10091242
Hormone-Independent Mouse Mammary Adenocarcinomas with Different Metastatic Potential Exhibit Different Metabolic Signatures
Abstract
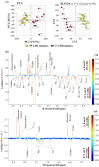
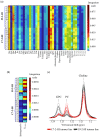
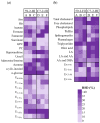
The metabolic characteristics of metastatic and non-metastatic breast carcinomas remain poorly studied. In this work, untargeted Nuclear Magnetic Resonance (NMR) metabolomics was used to compare two medroxyprogesterone acetate (MPA)-induced mammary carcinomas lines with different metastatic abilities. Different metabolic signatures distinguished the non-metastatic (59-2-HI) and the metastatic (C7-2-HI) lines, with glucose, amino acid metabolism, nucleotide metabolism and lipid metabolism as the major affected pathways. Non-metastatic tumours appeared to be characterised by: (a) reduced glycolysis and tricarboxylic acid cycle (TCA) activities, possibly resulting in slower NADH biosynthesis and reduced mitochondrial transport chain activity and ATP synthesis; (b) glutamate accumulation possibly related to reduced glutathione activity and reduced mTORC1 activity; and (c) a clear shift to lower phosphoscholine/glycerophosphocholine ratios and sphingomyelin levels. Within each tumour line, metabolic profiles also differed significantly between tumours (i.e., mice). Metastatic tumours exhibited marked inter-tumour changes in polar compounds, some suggesting different glycolytic capacities. Such tumours also showed larger intra-tumour variations in metabolites involved in nucleotide and cholesterol/fatty acid metabolism, in tandem with less changes in TCA and phospholipid metabolism, compared to non-metastatic tumours. This study shows the valuable contribution of untargeted NMR metabolomics to characterise tumour metabolism, thus opening enticing opportunities to find metabolic markers related to metastatic ability in endocrine breast cancer.
Keywords: NMR; endocrine breast cancer; hormone-independent growth; medroxyprogesterone acetate; metabolism; metabolomics; metastatic potential; murine models.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Five novel hormone-responsive cell lines derived from murine mammary ductal carcinomas: in vivo and in vitro effects of estrogens and progestins.Cancer Res. 2001 Jan 1;61(1):293-302. Cancer Res. 2001. PMID: 11196177
-
Transforming growth factor-beta activities in 'in vivo' lines of hormone-dependent and independent mammary adenocarcinomas induced by medroxyprogesterone acetate in BALB/c mice.Breast Cancer Res Treat. 1990 Jul;16(1):29-39. doi: 10.1007/BF01806573. Breast Cancer Res Treat. 1990. PMID: 2145045
-
Mammary adenocarcinomas induced by medroxyprogesterone acetate: hormone dependence and EGF receptors of BALB/c in vivo sublines.Int J Cancer. 1989 May 15;43(5):845-50. doi: 10.1002/ijc.2910430518. Int J Cancer. 1989. PMID: 2523869
-
The MPA mouse breast cancer model: evidence for a role of progesterone receptors in breast cancer.Endocr Relat Cancer. 2009 Jun;16(2):333-50. doi: 10.1677/ERC-08-0244. Epub 2009 Feb 3. Endocr Relat Cancer. 2009. PMID: 19190078 Review.
-
Hormone-dependent mammary tumors in mice and rats as a model for human breast cancer (review).Anticancer Res. 1983 Jul-Aug;3(4):273-81. Anticancer Res. 1983. PMID: 6309070 Review.
Cited by
-
Metabolic Adaptations in an Endocrine-Related Breast Cancer Mouse Model Unveil Potential Markers of Tumor Response to Hormonal Therapy.Front Oncol. 2022 Mar 1;12:786931. doi: 10.3389/fonc.2022.786931. eCollection 2022. Front Oncol. 2022. PMID: 35299741 Free PMC article.
-
Tumor Lipid Signatures Are Descriptive of Acquisition of Therapy Resistance in an Endocrine-Related Breast Cancer Mouse Model.J Proteome Res. 2024 Aug 2;23(8):2815-2829. doi: 10.1021/acs.jproteome.3c00382. Epub 2023 Jul 27. J Proteome Res. 2024. PMID: 37497607 Free PMC article.
References
-
- The World Health Organization Breast Cancer. [(accessed on 15 July 2020)]; Available online: http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/
-
- Haukaas T.H., Euceda L.R., Giskeødegård G.F., Lamichhane S., Krohn M., Jernström S., Aure M.R., Lingjærde O.C., Schlichting E., Garred Ø., et al. Metabolic clusters of breast cancer in relation to gene- and protein expression subtypes. Cancer Metab. 2016;4:12. doi: 10.1186/s40170-016-0152-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- UID/CTM/50011/2019 , POCI-01-0145-FEDER-007679,UID/BIM/04501/2019, UID/BIM/04501/2020 and POCI-01-0145-FEDER-007628/Fundação para a Ciência e a Tecnologia/International
- Infrastructure Project Nº 022161/Portuguese National NMR Network (PTNMR)/International
- POCI-01-0145-FEDER-028835/Integrated Programme CENTRO-01-0145-FEDER-000003/International
- ERC-2014-AdG - 669858/Fundação para a Ciência e a Tecnologia/International
- PhD grant (no grant number)/FCT-SPQ (Portuguese Chemical Society)/Periodic Table protocol/International
LinkOut - more resources
Full Text Sources
Miscellaneous

