Characterization of the Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family from Triticeae Species
- PMID: 32867204
- PMCID: PMC7504674
- DOI: 10.3390/ijms21176191
Characterization of the Heavy-Metal-Associated Isoprenylated Plant Protein (HIPP) Gene Family from Triticeae Species
Abstract
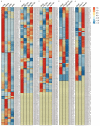
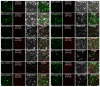
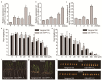
Heavy-metal-associated (HMA) isoprenylated plant proteins (HIPPs) only exist in vascular plants. They play important roles in responses to biotic/abiotic stresses, heavy-metal homeostasis, and detoxification. However, research on the distribution, diversification, and function of HIPPs in Triticeae species is limited. In this study, a total of 278 HIPPs were identified from a database from five Triticeae species, and 13 were cloned from Haynaldia villosa. These genes were classified into five groups by phylogenetic analysis. Most HIPPs had one HMA domain, while 51 from Clade I had two, and all HIPPs had good collinear relationships between species or subgenomes. In silico expression profiling revealed that 44 of the 114 wheat HIPPs were dominantly expressed in roots, 43 were upregulated under biotic stresses, and 29 were upregulated upon drought or heat treatment. Subcellular localization analysis of the cloned HIPPs from H. villosa showed that they were expressed on the plasma membrane. HIPP1-V was upregulated in H. villosa after Cd treatment, and transgenic wheat plants overexpressing HIPP1-V showed enhanced Cd tolerance, as shown by the recovery of seed-germination and root-growth inhibition by supplementary Cd. This research provides a genome-wide overview of the Triticeae HIPP genes and proved that HIPP1-V positively regulates Cd tolerance in common wheat.
Keywords: Cd tolerance; HIPP; Haynaldia villosa L.; gene family; subcellular localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Identification and Characterization of the EXO70 Gene Family in Polyploid Wheat and Related Species.Int J Mol Sci. 2018 Dec 24;20(1):60. doi: 10.3390/ijms20010060. Int J Mol Sci. 2018. PMID: 30586859 Free PMC article.
-
Heavy metal-associated isoprenylated plant protein (HIPP): characterization of a family of proteins exclusive to plants.FEBS J. 2013 Apr;280(7):1604-16. doi: 10.1111/febs.12159. Epub 2013 Feb 28. FEBS J. 2013. PMID: 23368984
-
Characterization of sucrose nonfermenting-1-related protein kinase 2 (SnRK2) gene family in Haynaldia villosa demonstrated SnRK2.9-V enhances drought and salt stress tolerance of common wheat.BMC Genomics. 2024 Feb 26;25(1):209. doi: 10.1186/s12864-024-10114-7. BMC Genomics. 2024. PMID: 38408894 Free PMC article.
-
Isolation and characterisation of cDNA encoding a wheat heavy metal-associated isoprenylated protein involved in stress responses.Plant Biol (Stuttg). 2015 Nov;17(6):1176-86. doi: 10.1111/plb.12344. Epub 2015 Sep 8. Plant Biol (Stuttg). 2015. PMID: 25951496
-
Heavy Metal-Associated Isoprenylated Plant Proteins (HIPPs) at Plasmodesmata: Exploring the Link between Localization and Function.Plants (Basel). 2023 Aug 21;12(16):3015. doi: 10.3390/plants12163015. Plants (Basel). 2023. PMID: 37631227 Free PMC article. Review.
Cited by
-
Dose-Dependent Physiological and Transcriptomic Responses of Lettuce (Lactuca sativa L.) to Copper Oxide Nanoparticles-Insights into the Phytotoxicity Mechanisms.Int J Mol Sci. 2021 Apr 1;22(7):3688. doi: 10.3390/ijms22073688. Int J Mol Sci. 2021. PMID: 33916236 Free PMC article.
-
The Effect of Cadmium on Plants in Terms of the Response of Gene Expression Level and Activity.Plants (Basel). 2023 Apr 30;12(9):1848. doi: 10.3390/plants12091848. Plants (Basel). 2023. PMID: 37176906 Free PMC article. Review.
-
Molecular Mechanisms Underlying Mimosa acutistipula Success in Amazonian Rehabilitating Minelands.Int J Environ Res Public Health. 2022 Nov 4;19(21):14441. doi: 10.3390/ijerph192114441. Int J Environ Res Public Health. 2022. PMID: 36361325 Free PMC article.
-
Overexpression of PavHIPP16 from Prunus avium enhances cold stress tolerance in transgenic tobacco.BMC Plant Biol. 2024 Jun 12;24(1):536. doi: 10.1186/s12870-024-05267-2. BMC Plant Biol. 2024. PMID: 38862890 Free PMC article.
-
Comprehensive Analysis of BrHMPs Reveals Potential Roles in Abiotic Stress Tolerance and Pollen-Stigma Interaction in Brassica rapa.Cells. 2023 Apr 6;12(7):1096. doi: 10.3390/cells12071096. Cells. 2023. PMID: 37048168 Free PMC article.
References
-
- Fang W., Wu P. Elevated selenium and other mineral element concentrations in soil and plant tissue in bone coal sites in Haoping area, Ziyang County, China. Plant Soil. 2004;261:135–146. doi: 10.1023/B:PLSO.0000035580.32406.e3. - DOI
MeSH terms
Substances
Grants and funding
- 2016YFD0101004/National Key Research and Development Program
- 2016YFD0102001-004/National Key Research and Development Program
- 91935304/National Natural Science Foundation of China (Major Program)
- 31661143005/International Cooperation and Exchange of the National Natural Science Foundation of China
- 31971943/National Natural Science Foundation of China
- 2015-Z41/'948' Project of Ministry of Agriculture
- BA2017138/Jiangsu Province for the transformation of scientific and technological achievements
- B08025/Program of Introducing Talents of Discipline to Universities
- PZCZ201706/Creation of Major New Agricultural Varieties in Jiangsu Province
- JATS [2019] 429/Jiangsu Agricultural Technology System (JATS)
- 2019BBF02022-04/Key Research and Development Major Project of Ningxia Autonomous Region
LinkOut - more resources
Full Text Sources

