The Role of Proteomics in Bacterial Response to Antibiotics
- PMID: 32867221
- PMCID: PMC7559545
- DOI: 10.3390/ph13090214
The Role of Proteomics in Bacterial Response to Antibiotics
Abstract
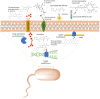
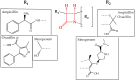
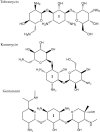
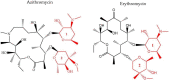
For many years, we have tried to use antibiotics to eliminate the persistence of pathogenic bacteria. However, these infectious agents can recover from antibiotic challenges through various mechanisms, including drug resistance and antibiotic tolerance, and continue to pose a global threat to human health. To design more efficient treatments against bacterial infections, detailed knowledge about the bacterial response to the commonly used antibiotics is required. Proteomics is a well-suited and powerful tool to study molecular response to antimicrobial compounds. Bacterial response profiling from system-level investigations could increase our understanding of bacterial adaptation, the mechanisms behind antibiotic resistance and tolerance development. In this review, we aim to provide an overview of bacterial response to the most common antibiotics with a focus on the identification of dynamic proteome responses, and through published studies, to elucidate the formation mechanism of resistant and tolerant bacterial phenotypes.
Keywords: antibiotic resistance; antibiotic tolerance; antibiotics; multi-drug resistant bacteria; pathogens; proteomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Proteomics in antibiotic resistance and tolerance research: Mapping the resistome and the tolerome of bacterial pathogens.Proteomics. 2022 Apr;22(8):e2100409. doi: 10.1002/pmic.202100409. Epub 2022 Mar 6. Proteomics. 2022. PMID: 35143120 Review.
-
Selective Proteomic Analysis of Antibiotic-Tolerant Cellular Subpopulations in Pseudomonas aeruginosa Biofilms.mBio. 2017 Oct 24;8(5):e01593-17. doi: 10.1128/mBio.01593-17. mBio. 2017. PMID: 29066549 Free PMC article.
-
The Neutrally Charged Diarylurea Compound PQ401 Kills Antibiotic-Resistant and Antibiotic-Tolerant Staphylococcus aureus.mBio. 2020 Jun 30;11(3):e01140-20. doi: 10.1128/mBio.01140-20. mBio. 2020. PMID: 32605985 Free PMC article.
-
Proteomics approach to understand bacterial antibiotic resistance strategies.Expert Rev Proteomics. 2019 Oct;16(10):829-839. doi: 10.1080/14789450.2019.1681978. Epub 2019 Oct 24. Expert Rev Proteomics. 2019. PMID: 31618606 Review.
-
Antibiotic resistant bacteria: current situation and treatment options to accelerate the development of a new antimicrobial arsenal.Expert Rev Anti Infect Ther. 2022 Aug;20(8):1095-1108. doi: 10.1080/14787210.2022.2078308. Epub 2022 May 31. Expert Rev Anti Infect Ther. 2022. PMID: 35576494 Review.
Cited by
-
New Insights into the Mechanism of Antibacterial Action of Synthetic Peptide Mo-CBP3-PepI against Klebsiella pneumoniae.Antibiotics (Basel). 2022 Dec 4;11(12):1753. doi: 10.3390/antibiotics11121753. Antibiotics (Basel). 2022. PMID: 36551410 Free PMC article.
-
Proteomic signatures of Staphylococcus aureus biofilm maturation on orthopaedic implants.Biofilm. 2025 May 27;9:100287. doi: 10.1016/j.bioflm.2025.100287. eCollection 2025 Jun. Biofilm. 2025. PMID: 40519940 Free PMC article.
-
Metaproteomics in the One Health framework for unraveling microbial effectors in microbiomes.Microbiome. 2025 May 23;13(1):134. doi: 10.1186/s40168-025-02119-5. Microbiome. 2025. PMID: 40410872 Free PMC article. Review.
-
A novel strategy to characterize the pattern of β-lactam antibiotic-induced drug resistance in Acinetobacter baumannii.Sci Rep. 2023 Jun 6;13(1):9177. doi: 10.1038/s41598-023-36475-9. Sci Rep. 2023. PMID: 37280269 Free PMC article.
-
XENOFOOD-An Autoclaved Feed Supplement Containing Autoclavable Antimicrobial Peptides-Exerts Anticoccidial GI Activity, and Causes Bursa Enlargement, but Has No Detectable Harmful Effects in Broiler Cockerels despite In Vitro Detectable Cytotoxicity on LHM Cells.Pathogens. 2023 Mar 14;12(3):458. doi: 10.3390/pathogens12030458. Pathogens. 2023. PMID: 36986380 Free PMC article.
References
-
- Antibiotic resistance threats in the United States 2019. [(accessed on 10 June 2020)]; Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-re....
-
- Cassini A., Högberg L.D., Plachouras D., Quattrocchi A., Hoxha A., Simonsen G.S., Colomb-Cotinat M., Kretzschmar M.E., Devleesschauwer B., Cecchini M., et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet. Infect. Dis. 2019;19:56–66. doi: 10.1016/S1473-3099(18)30605-4. - DOI - PMC - PubMed
-
- The Review on Antimicrobial Resistance. [(accessed on 10 June 2020)];:1–33. doi: 10.1016/j.jpha.2015.11.005. Available online: https://Amr-Review.Org/ - DOI
Publication types
LinkOut - more resources
Full Text Sources

