Potential Associations Among Alteration of Salivary miRNAs, Saliva Microbiome Structure, and Cognitive Impairments in Autistic Children
- PMID: 32867322
- PMCID: PMC7504581
- DOI: 10.3390/ijms21176203
Potential Associations Among Alteration of Salivary miRNAs, Saliva Microbiome Structure, and Cognitive Impairments in Autistic Children
Abstract
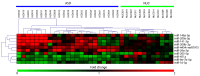
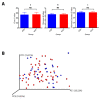
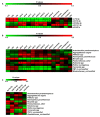
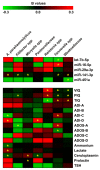
Recent evidence has demonstrated that salivary molecules, as well as bacterial populations, can be perturbed by several pathological conditions, including neuro-psychiatric diseases. This relationship between brain functionality and saliva composition could be exploited to unveil new pathological mechanisms of elusive diseases, such as Autistic Spectrum Disorder (ASD). We performed a combined approach of miRNA expression profiling by NanoString technology, followed by validation experiments in qPCR, and 16S rRNA microbiome analysis on saliva from 53 ASD and 27 neurologically unaffected control (NUC) children. MiR-29a-3p and miR-141-3p were upregulated, while miR-16-5p, let-7b-5p, and miR-451a were downregulated in ASD compared to NUCs. Microbiome analysis on the same subjects revealed that Rothia, Filifactor, Actinobacillus, Weeksellaceae, Ralstonia, Pasteurellaceae, and Aggregatibacter increased their abundance in ASD patients, while Tannerella, Moryella and TM7-3 decreased. Variations of both miRNAs and microbes were statistically associated to different neuropsychological scores related to anomalies in social interaction and communication. Among miRNA/bacteria associations, the most relevant was the negative correlation between salivary miR-141-3p expression and Tannerella abundance. MiRNA and microbiome dysregulations found in the saliva of ASD children are potentially associated with cognitive impairments of the subjects. Furthermore, a potential cross-talking between circulating miRNAs and resident bacteria could occur in saliva of ASD.
Keywords: ASD; Illumina; Nanostring; TaqMan assays; correlations; dysbiosis; microRNA; oral cavity; oral microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

