Predicting the most deleterious missense nsSNPs of the protein isoforms of the human HLA-G gene and in silico evaluation of their structural and functional consequences
- PMID: 32867672
- PMCID: PMC7457528
- DOI: 10.1186/s12863-020-00890-y
Predicting the most deleterious missense nsSNPs of the protein isoforms of the human HLA-G gene and in silico evaluation of their structural and functional consequences
Abstract
Background: The Human Leukocyte Antigen G (HLA-G) protein is an immune tolerogenic molecule with 7 isoforms. The change of expression level and some polymorphisms of the HLA-G gene are involved in various pathologies. Therefore, this study aimed to predict the most deleterious missense non-synonymous single nucleotide polymorphisms (nsSNPs) in HLA-G isoforms via in silico analyses and to examine structural and functional effects of the predicted nsSNPs on HLA-G isoforms.
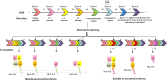
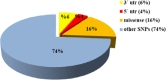
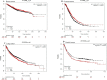
Results: Out of 301 reported SNPs in dbSNP, 35 missense SNPs in isoform 1, 35 missense SNPs in isoform 5, 8 missense SNPs in all membrane-bound HLA-G isoforms and 8 missense SNPs in all soluble HLA-G isoforms were predicted as deleterious by all eight servers (SIFT, PROVEAN, PolyPhen-2, I-Mutant 3.0, SNPs&GO, PhD-SNP, SNAP2, and MUpro). The Structural and functional effects of the predicted nsSNPs on HLA-G isoforms were determined by MutPred2 and HOPE servers, respectively. Consurf analyses showed that the majority of the predicted nsSNPs occur in conserved sites. I-TASSER and Chimera were used for modeling of the predicted nsSNPs. rs182801644 and rs771111444 were related to creating functional patterns in 5'UTR. 5 SNPs in 3'UTR of the HLA-G gene were predicted to affect the miRNA target sites. Kaplan-Meier analysis showed the HLA-G deregulation can serve as a prognostic marker for some cancers.
Conclusions: The implementation of in silico SNP prioritization methods provides a great framework for the recognition of functional SNPs. The results obtained from the current study would be called laboratory investigations.
Keywords: Deleterious SNPs; HLA-G gene; In silico analysis; Missense mutation; Structural and functional impact.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Group ISMW A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928. - PubMed
-
- Ding C, Jin S. High-throughput methods for SNP genotyping. In: Single Nucleotide Polymorphisms. Springer, Methods Mol Biol. 2009;578:245–54. https://link.springer.com/protocol/10.1007/978-1-60327-411-1_16. - DOI - PubMed
-
- Rajasekaran R, Doss CGP, Sudandiradoss C, Ramanathan K, Sethumadhavan R. In silico analysis of structural and functional consequences in p16INK4A by deleterious nsSNPs associated CDKN2A gene in malignant melanoma. Biochimie. 2008;90(10):1523–1529. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

