Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula
- PMID: 32867687
- PMCID: PMC7460785
- DOI: 10.1186/s12870-020-02619-6
Genome-wide study of C2H2 zinc finger gene family in Medicago truncatula
Abstract
Background: C2H2 zinc finger proteins (C2H2 ZFPs) play vital roles in shaping many aspects of plant growth and adaptation to the environment. Plant genomes harbor hundreds of C2H2 ZFPs, which compose one of the most important and largest transcription factor families in higher plants. Although the C2H2 ZFP gene family has been reported in several plant species, it has not been described in the model leguminous species Medicago truncatula.
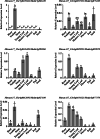
Results: In this study, we identified 218 C2H2 type ZFPs with 337 individual C2H2 motifs in M. truncatula. We showed that the high rate of local gene duplication has significantly contributed to the expansion of the C2H2 gene family in M. truncatula. The identified ZFPs exhibit high variation in motif arrangement and expression pattern, suggesting that the short C2H2 zinc finger motif has been adopted as a scaffold by numerous transcription factors with different functions to recognize cis-elements. By analyzing the public expression datasets and quantitative RT-PCR (qRT-PCR), we identified several C2H2 ZFPs that are specifically expressed in certain tissues, such as the nodule, seed, and flower.
Conclusion: Our genome-wide work revealed an expanded C2H2 ZFP gene family in an important legume M. truncatula, and provides new insights into the diversification and expansion of C2H2 ZFPs in higher plants.
Keywords: C2H2; EAR motif; Expression; Gene family; Local gene duplication; Zinc finger.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






Similar articles
-
Genome-Wide Identification of the Q-type C2H2 Transcription Factor Family in Alfalfa (Medicago sativa) and Expression Analysis under Different Abiotic Stresses.Genes (Basel). 2021 Nov 27;12(12):1906. doi: 10.3390/genes12121906. Genes (Basel). 2021. PMID: 34946855 Free PMC article.
-
Molecular characterization and expression analysis reveal the roles of Cys2/His2 zinc-finger transcription factors during flower development of Brassica rapa subsp. chinensis.Plant Mol Biol. 2020 Jan;102(1-2):123-141. doi: 10.1007/s11103-019-00935-6. Epub 2019 Nov 27. Plant Mol Biol. 2020. PMID: 31776846
-
Comprehensive genomic survey, structural classification and expression analysis of C2H2 zinc finger protein gene family in Brassica rapa L.PLoS One. 2019 May 6;14(5):e0216071. doi: 10.1371/journal.pone.0216071. eCollection 2019. PLoS One. 2019. PMID: 31059545 Free PMC article.
-
Genome-wide exploration of C2H2 zinc finger family in durum wheat (Triticum turgidum ssp. Durum): insights into the roles in biological processes especially stress response.Biometals. 2018 Dec;31(6):1019-1042. doi: 10.1007/s10534-018-0146-y. Epub 2018 Oct 4. Biometals. 2018. PMID: 30288657 Review.
-
Cys₂/His₂ Zinc-Finger Proteins in Transcriptional Regulation of Flower Development.Int J Mol Sci. 2018 Aug 31;19(9):2589. doi: 10.3390/ijms19092589. Int J Mol Sci. 2018. PMID: 30200325 Free PMC article. Review.
Cited by
-
Genome-wide analysis of the C2H2 zinc finger protein gene family and its response to salt stress in ginseng, Panax ginseng Meyer.Sci Rep. 2022 Jun 17;12(1):10165. doi: 10.1038/s41598-022-14357-w. Sci Rep. 2022. PMID: 35715520 Free PMC article.
-
Structures and biological functions of zinc finger proteins and their roles in hepatocellular carcinoma.Biomark Res. 2022 Jan 9;10(1):2. doi: 10.1186/s40364-021-00345-1. Biomark Res. 2022. PMID: 35000617 Free PMC article. Review.
-
De novo transcriptome assembly and analysis of gene expression in different tissues of moth bean (Vigna aconitifolia) (Jacq.) Marechal.BMC Plant Biol. 2022 Apr 15;22(1):198. doi: 10.1186/s12870-022-03583-z. BMC Plant Biol. 2022. PMID: 35428206 Free PMC article.
-
Genome-wide identification of the C2H2 zinc finger gene family and expression analysis under salt stress in sweetpotato.Front Plant Sci. 2023 Dec 13;14:1301848. doi: 10.3389/fpls.2023.1301848. eCollection 2023. Front Plant Sci. 2023. PMID: 38152142 Free PMC article.
-
Genome-wide identification and expression analysis of C2H2 zinc finger proteins in Chinese jujube (Ziziphus jujuba Mill.) in different fruit development stages and under different levels of water stress.PeerJ. 2024 Nov 29;12:e18455. doi: 10.7717/peerj.18455. eCollection 2024. PeerJ. 2024. PMID: 39624126 Free PMC article.
References
-
- Takatsuji H. Zinc-finger proteins: the classical zinc finger emerges in contemporary plant science. Plant Mol Biol. 1999;39(6):1073–1078. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

