MiR-199a-3p/5p participated in TGF-β and EGF induced EMT by targeting DUSP5/MAP3K11 in pterygium
- PMID: 32867783
- PMCID: PMC7461358
- DOI: 10.1186/s12967-020-02499-2
MiR-199a-3p/5p participated in TGF-β and EGF induced EMT by targeting DUSP5/MAP3K11 in pterygium
Abstract
Background: Recently, it has been reported that miRNA is involved in pterygium, however the exact underlying mechanism in pterygium is unrevealed and require further investigation.
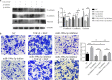
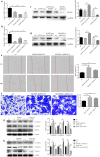
Methods: The differential expression of miRNA in pterygium was profiled using microarray and validated with quantitative real-time polymerase chain reaction (qRT-PCR). Human conjunctival epithelial cells (HCEs) were cultured and treated with transforming growth factor β (TGF-β) and epidermal growth factor (EGF) and transfected with miR-199a-3p/5p mimic and inhibitor. Markers of epithelial-mesenchymal transition (EMT) in HCEs were detected using western blot and immunohistochemistry. Cell migration ability was determined using wound healing and transwell assay, while apoptosis was determined by flow cytometry. The target genes of miR-199a were confirmed by the dual-luciferase reporter assay.
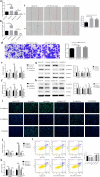
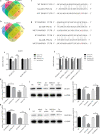
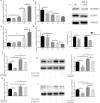
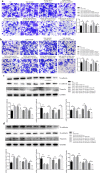
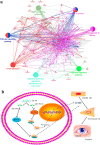
Results: TGF-β and EGF could induced EMT in HCEs and increase miR-199a-3p/5p but suppress target genes, DUSP5 and MAP3K11. With the occurrence of EMT, cell migration ability was enhanced, and apoptosis was impeded. Promoting miR-199a-3p/5p expression could induce EMT in HCEs without TGF-β and EGF, while suppressing miR-199a-3p/5p could inhibit EMT in TGF-β and EGF induced HCEs. In a word, TGF-β and EGF induced EMT could be regulated with miR-199a-3p/5p-DUSP5/MAP3K11 axes. The validated results in tissues showed that, compared with control conjunctival tissues, miR-199a-3p/5p were more overexpressed in pterygium, while DUSP5/MAP3K11 were lower expressed. In addition, bioinformatics analysis indicated the miR-199a-3p/5p-DUSP5/MAP3K11 was belong to MAPK signalling pathway.
Conclusions: TGF-β and EGF induce EMT of HCEs through miR-199a-3p/5p-DUSP5/MAP3K11 axes, which explains the pathogenesis of EMT in pterygium and may provide new targets for pterygium prevention and therapy.
Keywords: DUSP5; EMT; MAP3K11; Pterygium; miR-199a.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Identification of the circRNA-miRNA-mRNA Regulatory Network in Pterygium-Associated Conjunctival Epithelium.Biomed Res Int. 2022 Nov 8;2022:2673890. doi: 10.1155/2022/2673890. eCollection 2022. Biomed Res Int. 2022. PMID: 36398070 Free PMC article.
-
Overexpression of miR-199a-5p decreases esophageal cancer cell proliferation through repression of mitogen-activated protein kinase kinase kinase-11 (MAP3K11).Oncotarget. 2016 Feb 23;7(8):8756-70. doi: 10.18632/oncotarget.6752. Oncotarget. 2016. PMID: 26717044 Free PMC article.
-
MiR-199a-5p suppresses proliferation and invasion of human laryngeal cancer cells.Eur Rev Med Pharmacol Sci. 2020 Dec;24(23):12200-12207. doi: 10.26355/eurrev_202012_24010. Eur Rev Med Pharmacol Sci. 2020. PMID: 33336738
-
Non-coding RNA Activated by DNA Damage: Review of Its Roles in the Carcinogenesis.Front Cell Dev Biol. 2021 Aug 13;9:714787. doi: 10.3389/fcell.2021.714787. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34485302 Free PMC article. Review.
-
A review on the role of LINC01133 in cancers.Cancer Cell Int. 2022 Aug 30;22(1):270. doi: 10.1186/s12935-022-02690-z. Cancer Cell Int. 2022. PMID: 36042493 Free PMC article. Review.
Cited by
-
Talin1 Induces Epithelial-Mesenchymal Transition to Facilitate Endometrial Cell Migration and Invasion in Adenomyosis Under the Regulation of microRNA-145-5p.Reprod Sci. 2021 May;28(5):1523-1539. doi: 10.1007/s43032-020-00444-8. Epub 2021 Feb 4. Reprod Sci. 2021. PMID: 33537874
-
Non-viral gene therapy using RNA interference with PDGFR-α mediated epithelial-mesenchymal transformation for proliferative vitreoretinopathy.Mater Today Bio. 2023 Apr 11;20:100632. doi: 10.1016/j.mtbio.2023.100632. eCollection 2023 Jun. Mater Today Bio. 2023. PMID: 37122836 Free PMC article.
-
An Overview of the Role of MicroRNAs on Carcinogenesis: A Focus on Cell Cycle, Angiogenesis and Metastasis.Int J Mol Sci. 2023 Apr 14;24(8):7268. doi: 10.3390/ijms24087268. Int J Mol Sci. 2023. PMID: 37108432 Free PMC article. Review.
-
The inhibitory effect of Osthole on A549 lung adenocarcinoma cells and its biomarker.Sci Rep. 2025 Apr 15;15(1):12948. doi: 10.1038/s41598-025-97305-8. Sci Rep. 2025. PMID: 40234644 Free PMC article.
-
Identification of the circRNA-miRNA-mRNA Regulatory Network in Pterygium-Associated Conjunctival Epithelium.Biomed Res Int. 2022 Nov 8;2022:2673890. doi: 10.1155/2022/2673890. eCollection 2022. Biomed Res Int. 2022. PMID: 36398070 Free PMC article.
References
-
- Lucas RM, McMichael AJ, Armstrong BK, Smith WT. Estimating the global disease burden due to ultraviolet radiation exposure. Int J Epidemiol. 2008;37:654–667. - PubMed
-
- Todani A, Melki SA. Pterygium: current concepts in pathogenesis and treatment. Int Ophthalmol Clin. 2009;49:21–30. - PubMed
-
- Kocamis O, Bilgec M. Evaluation of the recurrence rate for pterygium treated with conjunctival autograft. Graefes Arch Clin Exp Ophthalmol. 2014;252:817–820. - PubMed
-
- Di Girolamo N. Signalling pathways activated by ultraviolet radiation: role in ocular and cutaneous health. Curr Pharm Des. 2010;16:1358–1375. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

