Influence of the amino-terminal sequence on the structure and function of HIV integrase
- PMID: 32867805
- PMCID: PMC7457537
- DOI: 10.1186/s12977-020-00537-x
Influence of the amino-terminal sequence on the structure and function of HIV integrase
Abstract
Background: Antiretroviral therapy (ART) can mitigate the morbidity and mortality caused by the human immunodeficiency virus (HIV). Successful development of ART can be accelerated by accurate structural and biochemical data on targets and their responses to inhibitors. One important ART target, HIV integrase (IN), has historically been studied in vitro in a modified form adapted to bacterial overexpression, with a methionine or a longer fusion protein sequence at the N-terminus. In contrast, IN present in viral particles is produced by proteolytic cleavage of the Pol polyprotein, which leaves a phenylalanine at the N-terminus (IN 1F). Inspection of available structures suggested that added residues on the N-terminus might disrupt proper protein folding and formation of multimeric complexes.
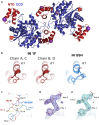
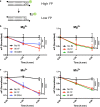
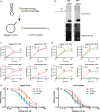
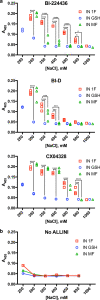
Results: We purified HIV-1 IN 1F1-212 and solved its structure at 2.4 Å resolution, which showed extension of an N-terminal helix compared to the published structure of IN1-212. Full-length IN 1F showed increased in vitro catalytic activity in assays of coupled joining of the two viral DNA ends compared to two IN variants containing additional N-terminal residues. IN 1F was also altered in its sensitivity to inhibitors, showing decreased sensitivity to the strand-transfer inhibitor raltegravir and increased sensitivity to allosteric integrase inhibitors. In solution, IN 1F exists as monomers and dimers, in contrast to other IN preparations which exist as higher-order oligomers.
Conclusions: The structural, biochemical, and biophysical characterization of IN 1F reveals the conformation of the native HIV-1 IN N-terminus and accompanying unique biochemical and biophysical properties. IN 1F thus represents an improved reagent for use in integration reactions in vitro and the development of antiretroviral agents.
Keywords: Biophysics; HIV; Integrases; Protein structure; Retroviridae; X-ray crystallography.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Characterization and structural analysis of HIV-1 integrase conservation.AIDS Rev. 2009 Jan-Mar;11(1):17-29. AIDS Rev. 2009. PMID: 19290031 Review.
-
HIV-1 integrase tetramers are the antiviral target of pyridine-based allosteric integrase inhibitors.Elife. 2019 May 23;8:e46344. doi: 10.7554/eLife.46344. Elife. 2019. PMID: 31120420 Free PMC article.
-
Consensus HIV-1 subtype A integrase and its raltegravir-resistant variants: design and characterization of the enzymatic properties.Biochimie. 2014 Jul;102:92-101. doi: 10.1016/j.biochi.2014.02.013. Epub 2014 Mar 2. Biochimie. 2014. PMID: 24594066
-
Allosteric inhibition of HIV-1 integrase activity.Curr Opin Chem Biol. 2013 Jun;17(3):339-45. doi: 10.1016/j.cbpa.2013.04.010. Epub 2013 May 3. Curr Opin Chem Biol. 2013. PMID: 23647983 Free PMC article. Review.
-
Crystal structure of the HIV-1 integrase core domain in complex with sucrose reveals details of an allosteric inhibitory binding site.FEBS Lett. 2010 Apr 16;584(8):1455-62. doi: 10.1016/j.febslet.2010.03.016. Epub 2010 Mar 16. FEBS Lett. 2010. PMID: 20227411
Cited by
-
Retroviral integrase: Structure, mechanism, and inhibition.Enzymes. 2021;50:249-300. doi: 10.1016/bs.enz.2021.06.007. Epub 2021 Aug 23. Enzymes. 2021. PMID: 34861940 Free PMC article.
-
Redondovirus Diversity and Evolution on Global, Individual, and Molecular Scales.J Virol. 2021 Oct 13;95(21):e0081721. doi: 10.1128/JVI.00817-21. Epub 2021 Aug 18. J Virol. 2021. PMID: 34406857 Free PMC article.
-
HIV-1 Integrase Assembles Multiple Species of Stable Synaptic Complex Intasomes That Are Active for Concerted DNA Integration In vitro.J Mol Biol. 2024 May 15;436(10):168557. doi: 10.1016/j.jmb.2024.168557. Epub 2024 Apr 4. J Mol Biol. 2024. PMID: 38582148 Free PMC article.
-
Structure of a HIV-1 IN-Allosteric inhibitor complex at 2.93 Å resolution: Routes to inhibitor optimization.PLoS Pathog. 2023 Mar 3;19(3):e1011097. doi: 10.1371/journal.ppat.1011097. eCollection 2023 Mar. PLoS Pathog. 2023. PMID: 36867659 Free PMC article.
References
-
- Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4(7):567–577. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-NS100081/NS/NINDS NIH HHS/United States
- T32 AI007632/AI/NIAID NIH HHS/United States
- R01-AI129661/National Institute of Allergy and Infectious Diseases/International
- U19-AI117950/National Institute of Allergy and Infectious Diseases/International
- UM1-AI126620/National Institute of Allergy and Infectious Diseases/International
LinkOut - more resources
Full Text Sources

