Treg-inducing capacity of genomic DNA of Bifidobacterium longum subsp. infantis
- PMID: 32867892
- PMCID: PMC8242987
- DOI: 10.2500/aap.2020.41.200064
Treg-inducing capacity of genomic DNA of Bifidobacterium longum subsp. infantis
Abstract
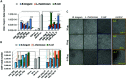
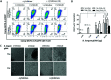
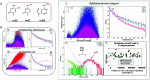
Background: Allergic and autoimmune diseases comprise a group of inflammatory disorders caused by aberrant immune responses in which CD25+ forkhead box P3-positive regulatory T cells (Treg) cells that normally suppress inflammatory events are often poorly functioning. This has stimulated an intensive investigative effort to find ways of increasing Tregs as a method of therapy for these conditions. Commensal microbiota known to have health benefits in humans include the lactic acid-producing, probiotic bacteria B. longum subsp. infantis and Lactobacillus rhamnosus. Mechanistically, several mechanisms have been proposed to explain how probiotics may favorably affect host immunity, including the induction of Tregs. Analysis of emerging data from several laboratories, including our own, suggest that DNA methylation may be an important determinant of immune reactivity responsible for Treg induction. Although methylated CpG moieties in normal mammalian DNA are both noninflammatory and lack immunogenicity, unmethylated CpGs, found largely in microbial DNA, are immunostimulatory and display proinflammatory properties. Objective: We hypothesize that microbiota with more DNA methylation may potentiate Treg induction to a greater degree than microbiota with a lower content of methylation. The purpose of the present study was to test this hypothesis by studying the methylation status of whole genomic DNA (gDNA) and the Treg-inducing capacity of purified gDNA in each of the probiotic bacteria B. longum subsp. infantis and L. rhamnosus, and a pathogenic Escherichia coli strain B. Results: We showed that gDNA from B. longum subsp. infantis is a potent Treg inducer that displays a dose-dependent response pattern at a dose threshold of 20 µg of gDNA. No similar Treg-inducing responses were observed with the gDNA from L. rhamnosus or E. coli. We identified a unique CpG methylated motif in the gDNA sequencing of B. longum subsp. infantis which was not found in L. rhamnosus or E. coli strain B. Conclusion: Although the literature indicates that both B. longum subsp. infantis and L. rhamnosus strains contribute to health, our data suggest that they do so by different mechanisms. Further, because of its small molecular size, low cost, ease of synthesis, and unique Treg-inducing feature, this methylated CpG oligodeoxynucleotide (ODN) from B. longum would offer many attractive features for an ideal novel therapeutic vaccine candidate for the treatment of immunologic diseases, such as the allergic and autoimmune disorders, in which Treg populations are diminished.
Conflict of interest statement
O.J. Lawless received patent 7,884,196 B2 “Vaccine Composition Comprising Methylated DNA and Immunomodulatory Motifs” was awarded on February 8, 2011. The remaining authors have no conflicts of interest to declare pertaining to this article
Figures

Similar articles
-
Dose-response studies of methylated and nonmethylated CpG ODNs from Bifidobacterium longum subsp. infantis for optimizing Treg cell stimulation.Allergy Asthma Proc. 2025 Mar 1;46(2):98-104. doi: 10.2500/aap.2025.46.250001. Allergy Asthma Proc. 2025. PMID: 40011992
-
Studies of methylated CpG ODN from Bifidobacterium longum subsp. infantis in a murine model: Implications for treatment of human allergic disease.Allergy Asthma Proc. 2025 Jan 1;46(1):e13-e23. doi: 10.2500/aap.2025.46.240100. Allergy Asthma Proc. 2025. PMID: 39741366
-
Methylated CpG ODNs from Bifidobacterium longum subsp. infantis Modulate Treg Induction and Suppress Allergic Response in a Murine Model.Int J Mol Sci. 2025 Jul 14;26(14):6755. doi: 10.3390/ijms26146755. Int J Mol Sci. 2025. PMID: 40725003 Free PMC article.
-
Impact of Bifidobacterium longum Subspecies infantis on Pediatric Gut Health and Nutrition: Current Evidence and Future Directions.Nutrients. 2024 Oct 16;16(20):3510. doi: 10.3390/nu16203510. Nutrients. 2024. PMID: 39458503 Free PMC article. Review.
-
Bifidobacterium longum Subspecies infantis (B. infantis) in Pediatric Nutrition: Current State of Knowledge.Nutrients. 2020 May 28;12(6):1581. doi: 10.3390/nu12061581. Nutrients. 2020. PMID: 32481558 Free PMC article. Review.
Cited by
-
Interconnection of the Gut-Skin Axis in NC/Nga Mouse with Atopic Dermatitis: Effects of the Three Types of Bifidobacterium bifidum CBT-BF3 (Probiotics, Postbiotics, and Cytosine-Phosphate-Guanine Oligodeoxynucleotide) on T Cell Differentiation and Gut Microbiota.Food Sci Anim Resour. 2024 Nov;44(6):1417-1439. doi: 10.5851/kosfa.2024.e100. Epub 2024 Nov 1. Food Sci Anim Resour. 2024. PMID: 39554831 Free PMC article.
-
Blood and guts: how the intestinal microbiome shapes hematopoiesis and treatment of hematologic disease.Blood. 2024 Apr 25;143(17):1689-1701. doi: 10.1182/blood.2023021174. Blood. 2024. PMID: 38364184 Free PMC article. Review.
-
LACpG10-HL Functions Effectively in Antibiotic-Free and Healthy Husbandry by Improving the Innate Immunity.Int J Mol Sci. 2022 Sep 28;23(19):11466. doi: 10.3390/ijms231911466. Int J Mol Sci. 2022. PMID: 36232768 Free PMC article.
-
Defining how microorganisms benefit human health.Microb Biotechnol. 2021 Jan;14(1):35-40. doi: 10.1111/1751-7915.13685. Epub 2020 Oct 25. Microb Biotechnol. 2021. PMID: 33099885 Free PMC article.
-
Allergen immunotherapy: From bench research to clinical application.Allergy Asthma Proc. 2020 Sep 1;41(5):311-313. doi: 10.2500/aap.2020.41.200070. Allergy Asthma Proc. 2020. PMID: 32867885 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

