Lysozyme Resistance in Clostridioides difficile Is Dependent on Two Peptidoglycan Deacetylases
- PMID: 32868404
- PMCID: PMC7585060
- DOI: 10.1128/JB.00421-20
Lysozyme Resistance in Clostridioides difficile Is Dependent on Two Peptidoglycan Deacetylases
Abstract
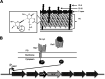
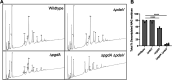
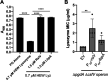
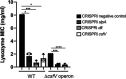
Clostridioides (Clostridium) difficile is a major cause of hospital-acquired infections leading to antibiotic-associated diarrhea. C. difficile exhibits a very high level of resistance to lysozyme. Bacteria commonly resist lysozyme through modification of the cell wall. In C. difficile, σV is required for lysozyme resistance, and σV is activated in response to lysozyme. Once activated, σV, encoded by csfV, directs transcription of genes necessary for lysozyme resistance. Here, we analyze the contribution of individual genes in the σV regulon to lysozyme resistance. Using CRISPR-Cas9-mediated mutagenesis we constructed in-frame deletions of single genes in the csfV operon. We find that pdaV, which encodes a peptidoglycan deacetylase, is partially responsible for lysozyme resistance. We then performed CRISPR inhibition (CRISPRi) to identify a second peptidoglycan deacetylase, encoded by pgdA, that is important for lysozyme resistance. Deletion of either pgdA or pdaV resulted in modest decreases in lysozyme resistance. However, deletion of both pgdA and pdaV resulted in a 1,000-fold decrease in lysozyme resistance. Further, muropeptide analysis revealed that loss of either PgdA or PdaV had modest effects on peptidoglycan deacetylation but that loss of both PgdA and PdaV resulted in almost complete loss of peptidoglycan deacetylation. This suggests that PgdA and PdaV are redundant peptidoglycan deacetylases. We also used CRISPRi to compare other lysozyme resistance mechanisms and conclude that peptidoglycan deacetylation is the major mechanism of lysozyme resistance in C. difficileIMPORTANCEClostridioides difficile is the leading cause of hospital-acquired diarrhea. C. difficile is highly resistant to lysozyme. We previously showed that the csfV operon is required for lysozyme resistance. Here, we used CRISPR-Cas9 mediated mutagenesis and CRISPRi knockdown to show that peptidoglycan deacetylation is necessary for lysozyme resistance and is the major lysozyme resistance mechanism in C. difficile We show that two peptidoglycan deacetylases in C. difficile are partially redundant and are required for lysozyme resistance. PgdA provides an intrinsic level of deacetylation, and PdaV, encoded by a part of the csfV operon, provides lysozyme-induced peptidoglycan deacetylation.
Keywords: cell envelope; gene expression; signal transduction; stress response; σ factors.
Copyright © 2020 American Society for Microbiology.
Figures
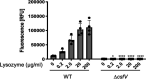


Similar articles
-
Peptidoglycan analysis reveals that synergistic deacetylase activity in vegetative Clostridium difficile impacts the host response.J Biol Chem. 2020 Dec 4;295(49):16785-16796. doi: 10.1074/jbc.RA119.012442. Epub 2020 Sep 25. J Biol Chem. 2020. PMID: 32978253 Free PMC article.
-
Clostridium difficile extracytoplasmic function σ factor σV regulates lysozyme resistance and is necessary for pathogenesis in the hamster model of infection.Infect Immun. 2014 Jun;82(6):2345-55. doi: 10.1128/IAI.01483-13. Epub 2014 Mar 24. Infect Immun. 2014. PMID: 24664503 Free PMC article.
-
Activation of the Extracytoplasmic Function σ Factor σV in Clostridioides difficile Requires Regulated Intramembrane Proteolysis of the Anti-σ Factor RsiV.mSphere. 2022 Apr 27;7(2):e0009222. doi: 10.1128/msphere.00092-22. Epub 2022 Mar 23. mSphere. 2022. PMID: 35317618 Free PMC article.
-
Activation of the extracytoplasmic function σ factor σV by lysozyme in Clostridioides difficile.Curr Opin Microbiol. 2022 Feb;65:162-166. doi: 10.1016/j.mib.2021.11.008. Epub 2021 Dec 8. Curr Opin Microbiol. 2022. PMID: 34894542 Free PMC article. Review.
-
Clostridioides difficile peptidoglycan modifications.Curr Opin Microbiol. 2022 Feb;65:156-161. doi: 10.1016/j.mib.2021.11.010. Epub 2021 Dec 6. Curr Opin Microbiol. 2022. PMID: 34883390 Review.
Cited by
-
Identification of Clostridioides difficile mutants with increased daptomycin resistance.J Bacteriol. 2024 Mar 21;206(3):e0036823. doi: 10.1128/jb.00368-23. Epub 2024 Feb 20. J Bacteriol. 2024. PMID: 38376203 Free PMC article.
-
Identification of a family of peptidoglycan transpeptidases reveals that Clostridioides difficile requires noncanonical cross-links for viability.Proc Natl Acad Sci U S A. 2024 Aug 20;121(34):e2408540121. doi: 10.1073/pnas.2408540121. Epub 2024 Aug 16. Proc Natl Acad Sci U S A. 2024. PMID: 39150786 Free PMC article.
-
Flagellar switch inverted repeat impacts flagellar invertibility and varies Clostridioides difficile RT027/MLST1 virulence.bioRxiv [Preprint]. 2024 Sep 24:2023.06.22.546185. doi: 10.1101/2023.06.22.546185. bioRxiv. 2024. PMID: 39386689 Free PMC article. Preprint.
-
Flagellar switch inverted repeats impact heterogeneity in flagellar gene expression and thus C. difficile RT027/MLST1 virulence.Cell Rep. 2025 Jun 24;44(6):115830. doi: 10.1016/j.celrep.2025.115830. Epub 2025 Jun 11. Cell Rep. 2025. PMID: 40504685 Free PMC article.
-
Virulence and genomic diversity among clinical isolates of ST1 (BI/NAP1/027) Clostridioides difficile.bioRxiv [Preprint]. 2023 Jan 12:2023.01.12.523823. doi: 10.1101/2023.01.12.523823. bioRxiv. 2023. Update in: Cell Rep. 2023 Aug 29;42(8):112861. doi: 10.1016/j.celrep.2023.112861. PMID: 36711955 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

