Plant expression of NifD protein variants resistant to mitochondrial degradation
- PMID: 32868448
- PMCID: PMC7502725
- DOI: 10.1073/pnas.2002365117
Plant expression of NifD protein variants resistant to mitochondrial degradation
Abstract
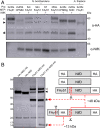
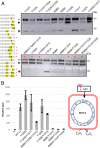
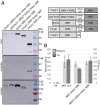
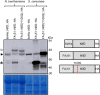
To engineer Mo-dependent nitrogenase function in plants, expression of the structural proteins NifD and NifK will be an absolute requirement. Although mitochondria have been established as a suitable eukaryotic environment for biosynthesis of oxygen-sensitive enzymes such as NifH, expression of NifD in this organelle has proven difficult due to cryptic NifD degradation. Here, we describe a solution to this problem. Using molecular and proteomic methods, we found NifD degradation to be a consequence of mitochondrial endoprotease activity at a specific motif within NifD. Focusing on this functionally sensitive region, we designed NifD variants comprising between one and three amino acid substitutions and distinguished several that were resistant to degradation when expressed in both plant and yeast mitochondria. Nitrogenase activity assays of these resistant variants in Escherichia coli identified a subset that retained function, including a single amino acid variant (Y100Q). We found that other naturally occurring NifD proteins containing alternate amino acids at the Y100 position were also less susceptible to degradation. The Y100Q variant also enabled expression of a NifD(Y100Q)-linker-NifK translational polyprotein in plant mitochondria, confirmed by identification of the polyprotein in the soluble fraction of plant extracts. The NifD(Y100Q)-linker-NifK retained function in bacterial nitrogenase assays, demonstrating that this polyprotein permits expression of NifD and NifK in a defined stoichiometry supportive of activity. Our results exemplify how protein design can overcome impediments encountered when expressing synthetic proteins in novel environments. Specifically, these findings outline our progress toward the assembly of the catalytic unit of nitrogenase within mitochondria.
Keywords: metabolic engineering; mitochondria; nitrogenase; protein engineering; synthetic biology.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

