The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development
- PMID: 32868762
- PMCID: PMC7459115
- DOI: 10.1038/s41467-020-18231-z
The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development
Erratum in
-
Author Correction: The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development.Nat Commun. 2022 Jan 11;13(1):332. doi: 10.1038/s41467-021-27676-9. Nat Commun. 2022. PMID: 35017490 Free PMC article. No abstract available.
Abstract
The hypothalamus is a central regulator of many innate behaviors essential for survival, but the molecular mechanisms controlling hypothalamic patterning and cell fate specification are poorly understood. To identify genes that control hypothalamic development, we have used single-cell RNA sequencing (scRNA-Seq) to profile mouse hypothalamic gene expression across 12 developmental time points between embryonic day 10 and postnatal day 45. This identified genes that delineated clear developmental trajectories for all major hypothalamic cell types, and readily distinguished major regional subdivisions of the developing hypothalamus. By using our developmental dataset, we were able to rapidly annotate previously unidentified clusters from existing scRNA-Seq datasets collected during development and to identify the developmental origins of major neuronal populations of the ventromedial hypothalamus. We further show that our approach can rapidly and comprehensively characterize mutants that have altered hypothalamic patterning, identifying Nkx2.1 as a negative regulator of prethalamic identity. These data serve as a resource for further studies of hypothalamic development, physiology, and dysfunction.
Conflict of interest statement
The authors declare no competing interests.
Figures
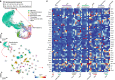


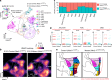
References
-
- Kent MA, Peters RH. Effects of ventromedial hypothalamic lesions on hunger-motivated behavior in rats. J. Comp. Physiol. Psychol. 1973;83:92–97. - PubMed
-
- Kruk MR, et al. Discriminant analysis of the localization of aggression-inducing electrode placements in the hypothalamus of male rats. Brain Res. 1983;260:61–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

