This is a preprint.
Genome-wide CRISPR screen reveals host genes that regulate SARS-CoV-2 infection
- PMID: 32869025
- PMCID: PMC7457610
- DOI: 10.1101/2020.06.16.155101
Genome-wide CRISPR screen reveals host genes that regulate SARS-CoV-2 infection
Update in
-
Genome-wide CRISPR Screens Reveal Host Factors Critical for SARS-CoV-2 Infection.Cell. 2021 Jan 7;184(1):76-91.e13. doi: 10.1016/j.cell.2020.10.028. Epub 2020 Oct 20. Cell. 2021. PMID: 33147444 Free PMC article.
Abstract
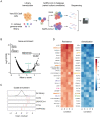
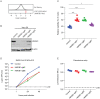
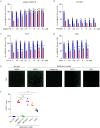
Identification of host genes essential for SARS-CoV-2 infection may reveal novel therapeutic targets and inform our understanding of COVID-19 pathogenesis. Here we performed a genome-wide CRISPR screen with SARS-CoV-2 and identified known SARS-CoV-2 host factors including the receptor ACE2 and protease Cathepsin L. We additionally discovered novel pro-viral genes and pathways including the SWI/SNF chromatin remodeling complex and key components of the TGF-β signaling pathway. Small molecule inhibitors of these pathways prevented SARS-CoV-2-induced cell death. We also revealed that the alarmin HMGB1 is critical for SARS-CoV-2 replication. In contrast, loss of the histone H3.3 chaperone complex sensitized cells to virus-induced death. Together this study reveals potential therapeutic targets for SARS-CoV-2 and highlights host genes that may regulate COVID-19 pathogenesis.
Conflict of interest statement
Competing interests
Yale University (CBW) has a patent pending related to this work entitled: “Compounds and Compositions for Treating, Ameliorating, and/or Preventing SARS-CoV-2 Infection and/or Complications Thereof.” Yale University has committed to rapidly executable non-exclusive royalty-free licenses to intellectual property rights for the purpose of making and distributing products to prevent, diagnose and treat COVID-19 infection during the pandemic and for a short period thereafter. JGD consults for Foghorn Therapeutics, Maze Therapeutics, Merck, Agios, and Pfizer; JGD consults for and has equity in Tango Therapeutics.
Figures


References
-
- Guan Y., Zheng B. J., He Y. Q., Liu X. L., Zhuang Z. X., Cheung C. L., Luo S. W., Li P. H., Zhang L. J., Guan Y. J., Butt K. M., Wong K. L., Chan K. W., Lim W., Shortridge K. F., Yuen K. Y., Peiris J. S., Poon L. L., Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 302, 276–278 (2003). - PubMed
Publication types
Grants and funding
- R01 AI123449/AI/NIAID NIH HHS/United States
- T32 AI007019/AI/NIAID NIH HHS/United States
- T32 GM007223/GM/NIGMS NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- F30 HL149151/HL/NHLBI NIH HHS/United States
- R21 AI157835/AI/NIAID NIH HHS/United States
- T32 GM007205/GM/NIGMS NIH HHS/United States
- P50 CA121974/CA/NCI NIH HHS/United States
- K08 AI128043/AI/NIAID NIH HHS/United States
- R01 AI087925/AI/NIAID NIH HHS/United States
- F31 DA054739/DA/NIDA NIH HHS/United States
- U19 AI133524/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
