Biomarkers in Breast Cancer: An Integrated Analysis of Comprehensive Genomic Profiling and PD-L1 Immunohistochemistry Biomarkers in 312 Patients with Breast Cancer
- PMID: 32869930
- PMCID: PMC7648336
- DOI: 10.1634/theoncologist.2020-0449
Biomarkers in Breast Cancer: An Integrated Analysis of Comprehensive Genomic Profiling and PD-L1 Immunohistochemistry Biomarkers in 312 Patients with Breast Cancer
Abstract
Background: We examined the current biomarker landscape in breast cancer when programmed death-ligand 1 (PD-L1) testing is integrated with comprehensive genomic profiling (CGP).
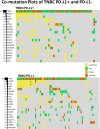
Material and methods: We analyzed data from samples of 312 consecutive patients with breast carcinoma tested with both CGP and PD-L1 (SP142) immunohistochemistry (IHC) during routine clinical care. These samples were stratified into hormone receptor positive (HR+)/human epidermal growth factor receptor negative (HER2-; n = 159), HER2-positive (n = 32), and triple-negative breast cancer (TNBC) cohorts (n = 121).
Results: We found that in the TNBC cohort, 43% (52/121) were immunocyte PD-L1-positive, and in the HR+/HER2- cohort, 30% (48/159) had PIK3CA companion diagnostics mutations, and hence were potentially eligible for atezolizumab plus nab-paclitaxel or alpelisib plus fulvestrant, respectively. Of the remaining 212 patients, 10.4% (22/212) had a BRCA1/2 mutation, which, if confirmed by germline testing, would allow olaparib plus talazoparib therapy. Of the remaining 190 patients, 169 (88.9%) were positive for another therapy-associated marker or a marker that would potentially qualify the patient for a clinical trial. In addition, we examined the relationship between immunocyte PD-L1 positivity and different tumor mutation burden (TMB) cutoffs and found that when a TMB cutoff of ≥9 mutations per Mb was applied (cutoff determined based on prior publication), 11.6% (14/121) patients were TMB ≥9 mutations/Mb and of these, TMB ≥9 mutations per Mb, 71.4% (10/14) were also positive for PD-L1 IHC.
Conclusion: Our integrated PD-L1 and CGP methodology identified 32% of the tested patients as potentially eligible for at least one of the two new Food and Drug Administration approved therapies, atezolizumab or alpelisib, and an additional 61.2% (191/312) had other biomarker-guided potential therapeutic options.
Implications for practice: This integrated programmed death-ligand 1 immunohistochemistry and comprehensive genomic profiling methodology identified 32% of the tested patients as eligible for at least one of the two new Food and Drug Administration-approved therapies, atezolizumab or alpelisib, and an additional 61.2% (191/312) had other biomarker-guided potential therapeutic options. These findings suggest new research opportunities to evaluate the predictive utility of other commonly seen PIK3CA mutations in hormone receptor-positive breast cancers and to standardize tumor mutation burden cutoffs to evaluate its potentially predictive role in triple-negative breast cancer.
Keywords: Biomarkers; Breast carcinoma; Comprehensive genomic profiling; PD-L1 immunohistochemistry.
© AlphaMed Press 2020.
Conflict of interest statement
Figures




References
-
- American Cancer Society . Facts & Figures 2019. Atlanta, GA: American Cancer Society; 2019.
-
- Duffy M, Harbeck N, Nap M et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM). Eur J Cancer 2017;75:284–298. - PubMed
-
- Food and Drug Administration . List of cleared or approved companion diagnostic devices (in vitro and imaging tools). Available at https://www.fda.gov/medical-devices/vitro-diagnostics/list-cleared-or-ap.... Accessed September 27, 2019.
-
- Schmid P, Adams S, Rugo H et al. Atezolizumab and nab‐paclitaxel in advanced triple‐negative breast cancer. N Engl J Med 2019;380:1929–1940. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

