A simple protein-based surrogate neutralization assay for SARS-CoV-2
- PMID: 32870820
- PMCID: PMC7566699
- DOI: 10.1172/jci.insight.142362
A simple protein-based surrogate neutralization assay for SARS-CoV-2
Abstract
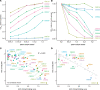
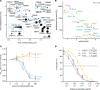
Most of the patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mount a humoral immune response to the virus within a few weeks of infection, but the duration of this response and how it correlates with clinical outcomes has not been completely characterized. Of particular importance is the identification of immune correlates of infection that would support public health decision-making on treatment approaches, vaccination strategies, and convalescent plasma therapy. While ELISA-based assays to detect and quantitate antibodies to SARS-CoV-2 in patient samples have been developed, the detection of neutralizing antibodies typically requires more demanding cell-based viral assays. Here, we present a safe and efficient protein-based assay for the detection of serum and plasma antibodies that block the interaction of the SARS-CoV-2 spike protein receptor binding domain (RBD) with its receptor, angiotensin-converting enzyme 2 (ACE2). The assay serves as a surrogate neutralization assay and is performed on the same platform and in parallel with an ELISA for the detection of antibodies against the RBD, enabling a direct comparison. The results obtained with our assay correlate with those of 2 viral-based assays, a plaque reduction neutralization test (PRNT) that uses live SARS-CoV-2 virus and a spike pseudotyped viral vector-based assay.
Keywords: Adaptive immunity; Antigen; Immunoglobulins; Infectious disease.
Conflict of interest statement
Figures

References
-
- Corti D, Lanzavecchia A. Broadly neutralizing antiviral antibodies. Annu Rev Immunol. 2013;31:705–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

