Col11a1a Expression Is Required for Zebrafish Development
- PMID: 32872105
- PMCID: PMC7558312
- DOI: 10.3390/jdb8030016
Col11a1a Expression Is Required for Zebrafish Development
Abstract
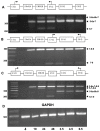
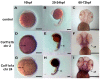
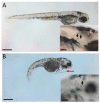
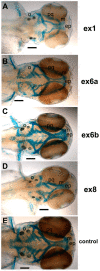
The autosomal dominant chondrodystrophies, the Stickler type 2 and Marshall syndromes, are characterized by facial abnormalities, vision deficits, hearing loss, and articular joint issues resulting from mutations in COL11A1. Zebrafish carry two copies of the Col11a1 gene, designated Col11a1a and Col11a1b. Col11a1a is located on zebrafish chromosome 24 and Col11a1b is located on zebrafish chromosome 2. Expression patterns are distinct for Col11a1a and Col11a1b and Col11a1a is most similar to COL11A1 that is responsible for human autosomal chondrodystrophies and the gene responsible for changes in the chondrodystrophic mouse model cho/cho. We investigated the function of Col11a1a in craniofacial and axial skeletal development in zebrafish using a knockdown approach. Knockdown revealed abnormalities in Meckel's cartilage, the otoliths, and overall body length. Similar phenotypes were observed using a CRISPR/Cas9 gene-editing approach, although the CRISPR/Cas9 effect was more severe compared to the transient effect of the antisense morpholino oligonucleotide treatment. The results of this study provide evidence that the zebrafish gene for Col11a1a is required for normal development and has similar functions to the mammalian COL11A1 gene. Due to its transparency, external fertilization, the Col11a1a knockdown, and knockout zebrafish model systems can, therefore, contribute to filling the gap in knowledge about early events during vertebrate skeletal development that are not as tenable in mammalian model systems and help us understand Col11a1-related early developmental events.
Keywords: Col11a1a; Marshall syndrome; Stickler type 2 syndrome; alternative splicing; collagen; fibrochondrogenesis; minor fibrillar collagen; zebrafish.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Authentication of a novel antibody to zebrafish collagen type XI alpha 1 chain (Col11a1a).BMC Res Notes. 2021 Sep 15;14(1):359. doi: 10.1186/s13104-021-05770-x. BMC Res Notes. 2021. PMID: 34526111 Free PMC article.
-
The Shape of the Jaw-Zebrafish Col11a1a Regulates Meckel's Cartilage Morphogenesis and Mineralization.J Dev Biol. 2022 Sep 22;10(4):40. doi: 10.3390/jdb10040040. J Dev Biol. 2022. PMID: 36278545 Free PMC article.
-
Craniofacial cartilage morphogenesis requires zebrafish col11a1 activity.Matrix Biol. 2009 Oct;28(8):490-502. doi: 10.1016/j.matbio.2009.07.004. Epub 2009 Jul 26. Matrix Biol. 2009. PMID: 19638309
-
Enhancing Understanding of the Visual Cycle by Applying CRISPR/Cas9 Gene Editing in Zebrafish.Front Cell Dev Biol. 2018 Apr 11;6:37. doi: 10.3389/fcell.2018.00037. eCollection 2018. Front Cell Dev Biol. 2018. PMID: 29696141 Free PMC article. Review.
-
Hearing Loss in Stickler Syndrome: An Update.Genes (Basel). 2022 Sep 1;13(9):1571. doi: 10.3390/genes13091571. Genes (Basel). 2022. PMID: 36140739 Free PMC article. Review.
Cited by
-
Authentication of a novel antibody to zebrafish collagen type XI alpha 1 chain (Col11a1a).BMC Res Notes. 2021 Sep 15;14(1):359. doi: 10.1186/s13104-021-05770-x. BMC Res Notes. 2021. PMID: 34526111 Free PMC article.
-
Early stress exposure on zebrafish development: effects on survival, malformations and molecular alterations.Fish Physiol Biochem. 2024 Aug;50(4):1545-1562. doi: 10.1007/s10695-024-01355-0. Epub 2024 May 14. Fish Physiol Biochem. 2024. PMID: 38743196 Free PMC article.
-
The Shape of the Jaw-Zebrafish Col11a1a Regulates Meckel's Cartilage Morphogenesis and Mineralization.J Dev Biol. 2022 Sep 22;10(4):40. doi: 10.3390/jdb10040040. J Dev Biol. 2022. PMID: 36278545 Free PMC article.
References
-
- Hall B.K., Miyake T. Divide, accumulate, differentiate: Cell condensation in skeletal development revisited. Int. J. Dev. Biol. 1995;39:881–893. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

