The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing
- PMID: 32873578
- PMCID: PMC7462061
- DOI: 10.1101/gad.339986.120
The nuclear cap-binding complex as choreographer of gene transcription and pre-mRNA processing
Abstract
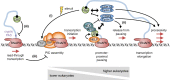
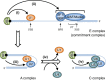
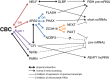
The largely nuclear cap-binding complex (CBC) binds to the 5' caps of RNA polymerase II (RNAPII)-synthesized transcripts and serves as a dynamic interaction platform for a myriad of RNA processing factors that regulate gene expression. While influence of the CBC can extend into the cytoplasm, here we review the roles of the CBC in the nucleus, with a focus on protein-coding genes. We discuss differences between CBC function in yeast and mammals, covering the steps of transcription initiation, release of RNAPII from pausing, transcription elongation, cotranscriptional pre-mRNA splicing, transcription termination, and consequences of spurious transcription. We describe parameters known to control the binding of generic or gene-specific cofactors that regulate CBC activities depending on the process(es) targeted, illustrating how the CBC is an ever-changing choreographer of gene expression.
Keywords: CBC; CBP20; CBP80; RNA 3′ end formation; alternative splicing; pre-mRNA splicing; promoter-proximal pausing; transcription elongation; transcription initiation; transcription preinitiation complex; transcription termination.
© 2020 Rambout and Maquat; Published by Cold Spring Harbor Laboratory Press.
Figures
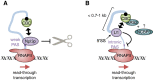

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
