Evolutionary Variation in MADS Box Dimerization Affects Floral Development and Protein Abundance in Maize
- PMID: 32873631
- PMCID: PMC7610293
- DOI: 10.1105/tpc.20.00300
Evolutionary Variation in MADS Box Dimerization Affects Floral Development and Protein Abundance in Maize
Abstract
Interactions between MADS box transcription factors are critical in the regulation of floral development, and shifting MADS box protein-protein interactions are predicted to have influenced floral evolution. However, precisely how evolutionary variation in protein-protein interactions affects MADS box protein function remains unknown. To assess the impact of changing MADS box protein-protein interactions on transcription factor function, we turned to the grasses, where interactions between B-class MADS box proteins vary. We tested the functional consequences of this evolutionary variability using maize (Zea mays) as an experimental system. We found that differential B-class dimerization was associated with subtle, quantitative differences in stamen shape. In contrast, differential dimerization resulted in large-scale changes to downstream gene expression. Differential dimerization also affected B-class complex composition and abundance, independent of transcript levels. This indicates that differential B-class dimerization affects protein degradation, revealing an important consequence for evolutionary variability in MADS box interactions. Our results highlight complexity in the evolution of developmental gene networks: changing protein-protein interactions could affect not only the composition of transcription factor complexes but also their degradation and persistence in developing flowers. Our results also show how coding change in a pleiotropic master regulator could have small, quantitative effects on development.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures

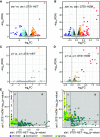
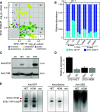


Comment in
-
Fine Tuning Floral Morphology: MADS-Box Protein Complex Formation in Maize.Plant Cell. 2020 Nov;32(11):3376-3377. doi: 10.1105/tpc.20.00818. Epub 2020 Oct 1. Plant Cell. 2020. PMID: 33004615 Free PMC article. No abstract available.
References
-
- Abraham-Juárez M.J.(2019). Western blot in maize. Bio-101 e3257.
-
- Ambrose B.A., Lerner D.R., Ciceri P., Padilla C.M., Yanofsky M.F., Schmidt R.J.(2000). Molecular and genetic analyses of the silky1 gene reveal conservation in floral organ specification between eudicots and monocots. Mol. Cell 5: 569–579. - PubMed
-
- Angenent G.C., Immink R.G.H.(2009). Combinatorial action of petunia MADS box genes and their protein products In Petunia: Evolutionary, Developmental and Physiological Genetics, Gerats T., and Strommer J., eds (New York: Springer; ), pp. 225–245.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

