Analysis of muntjac deer genome and chromatin architecture reveals rapid karyotype evolution
- PMID: 32873878
- PMCID: PMC7463020
- DOI: 10.1038/s42003-020-1096-9
Analysis of muntjac deer genome and chromatin architecture reveals rapid karyotype evolution
Abstract
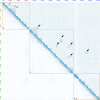
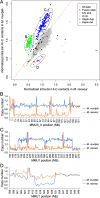
Closely related muntjac deer show striking karyotype differences. Here we describe chromosome-scale genome assemblies for Chinese and Indian muntjacs, Muntiacus reevesi (2n = 46) and Muntiacus muntjak vaginalis (2n = 6/7), and analyze their evolution and architecture. The genomes show extensive collinearity with each other and with other deer and cattle. We identified numerous fusion events unique to and shared by muntjacs relative to the cervid ancestor, confirming many cytogenetic observations with genome sequence. One of these M. muntjak fusions reversed an earlier fission in the cervid lineage. Comparative Hi-C analysis showed that the chromosome fusions on the M. muntjak lineage altered long-range, three-dimensional chromosome organization relative to M. reevesi in interphase nuclei including A/B compartment structure. This reshaping of multi-megabase contacts occurred without notable change in local chromatin compaction, even near fusion sites. A few genes involved in chromosome maintenance show evidence for rapid evolution, possibly associated with the dramatic changes in karyotype.
Conflict of interest statement
D.S.R. is a member of the Scientific Advisory Board of and a minor shareholder in Dovetail Genomics, which developed the Hi-C library preparation kit used in this study and performed quality control analyses on the Hi-C libraries. The remaining authors declare no competing interests.
Figures

References
-
- Wurster DH, Benirschke K. Indian muntjac, Muntiacus muntjak: a deer with a low diploid chromosome number. Science. 1970;168:1364–1366. - PubMed
-
- Wurster DH, Benirschke K. Chromosome studies in some deer, the springbok, and the pronghorn, with notes on placentation in deer. Cytologia. 1967;32:273–285. - PubMed
-
- Chi J, et al. New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis) Cytogenet. Genome Res. 2005;108:310–316. - PubMed
-
- Hsu TC, Pathak S, Chen TR. The possibility of latent centromeres and a proposed nomenclature system for total chromosome and whole arm translocations. Cytogenet. Cell Genet. 1975;15:41–49. - PubMed
-
- Liming S, Yingying Y, Xingsheng D. Comparative cytogenetic studies on the red muntjac, Chinese muntjac, and their F1 hybrids. Cytogenet. Genome Res. 1980;26:22–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA196884/CA/NCI NIH HHS/United States
- T32GM007127/GM/NIGMS NIH HHS/United States
- T32 HG000047/HG/NHGRI NIH HHS/United States
- S10OD018174/OD/NIH HHS/United States
- R01CA196884/CA/NCI NIH HHS/United States
- S10OD010786/OD/NIH HHS/United States
- S10 OD018174/OD/NIH HHS/United States
- R01 HD080708/HD/NICHD NIH HHS/United States
- S10 OD010786/OD/NIH HHS/United States
- T32 GM007127/GM/NIGMS NIH HHS/United States
- T32 GM066698/GM/NIGMS NIH HHS/United States
- R01GM086321/GM/NIGMS NIH HHS/United States
- T32HG000047/HG/NHGRI NIH HHS/United States
- R01 GM086321/GM/NIGMS NIH HHS/United States
- R01HD080708/NH/NIH HHS/United States
- T32GM066698/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources

