Identification of potential biomarkers and candidate small molecule drugs in glioblastoma
- PMID: 32874133
- PMCID: PMC7455906
- DOI: 10.1186/s12935-020-01515-1
Identification of potential biomarkers and candidate small molecule drugs in glioblastoma
Abstract
Background and aims: Glioblastoma (GBM) is a common and aggressive primary brain tumor, and the prognosis for GBM patients remains poor. This study aimed to identify the key genes associated with the development of GBM and provide new diagnostic and therapies for GBM.
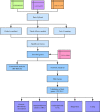
Methods: Three microarray datasets (GSE111260, GSE103227, and GSE104267) were selected from Gene Expression Omnibus (GEO) database for integrated analysis. The differential expressed genes (DEGs) between GBM and normal tissues were identified. Then, prognosis-related DEGs were screened by survival analysis, followed by functional enrichment analysis. The protein-protein interaction (PPI) network was constructed to explore the hub genes associated with GBM. The mRNA and protein expression levels of hub genes were respectively validated in silico using The Cancer Genome Atlas (TCGA) and Human Protein Atlas (HPA) databases. Subsequently, the small molecule drugs of GBM were predicted by using Connectivity Map (CMAP) database.
Results: A total of 78 prognosis-related DEGs were identified, of which10 hub genes with higher degree were obtained by PPI analysis. The mRNA expression and protein expression levels of CETN2, MKI67, ARL13B, and SETDB1 were overexpressed in GBM tissues, while the expression levels of CALN1, ELAVL3, ADCY3, SYN2, SLC12A5, and SOD1 were down-regulated in GBM tissues. Additionally, these genes were significantly associated with the prognosis of GBM. We eventually predicted the 10 most vital small molecule drugs, which potentially imitate or reverse GBM carcinogenic status. Cycloserine and 11-deoxy-16,16-dimethylprostaglandin E2 might be considered as potential therapeutic drugs of GBM.
Conclusions: Our study provided 10 key genes for diagnosis, prognosis, and therapy for GBM. These findings might contribute to a better comprehension of molecular mechanisms of GBM development, and provide new perspective for further GBM research. However, specific regulatory mechanism of these genes needed further elaboration.
Keywords: Differentially expressed genes; Glioblastoma; Hub genes; Prognosis; Small molecular drugs.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures






References
-
- Lukas RV, Rodon J, Becker K, Wong ET, Shih K, Touat M, et al. Clinical activity and safety of atezolizumab in patients with recurrent glioblastoma. J Neurooncol. 2018;140(2):317–328. - PubMed
-
- Alexander BM, Cloughesy TF. Adult glioblastoma. J Clin Oncol. 2017;35(21):2402–2409. - PubMed
-
- Delgado-López P, Corrales-García E. Survival in glioblastoma: a review on the impact of treatment modalities. Clin Transl Oncol. 2016;18(11):1062–1071. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

