Engineering Pseudomonas putida KT2440 for the production of isobutanol
- PMID: 32874178
- PMCID: PMC7447888
- DOI: 10.1002/elsc.201900151
Engineering Pseudomonas putida KT2440 for the production of isobutanol
Abstract
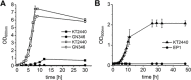
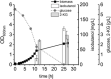
We engineered P. putida for the production of isobutanol from glucose by preventing product and precursor degradation, inactivation of the soluble transhydrogenase SthA, overexpression of the native ilvC and ilvD genes, and implementation of the feedback-resistant acetolactate synthase AlsS from Bacillus subtilis, ketoacid decarboxylase KivD from Lactococcus lactis, and aldehyde dehydrogenase YqhD from Escherichia coli. The resulting strain P. putida Iso2 produced isobutanol with a substrate specific product yield (Y Iso/S) of 22 ± 2 mg per gram of glucose under aerobic conditions. Furthermore, we identified the ketoacid decarboxylase from Carnobacterium maltaromaticum to be a suitable alternative for isobutanol production, since replacement of kivD from L. lactis in P. putida Iso2 by the variant from C. maltaromaticum yielded an identical YIso/S. Although P. putida is regarded as obligate aerobic, we show that under oxygen deprivation conditions this bacterium does not grow, remains metabolically active, and that engineered producer strains secreted isobutanol also under the non-growing conditions.
Keywords: Pseudomonas putida; isobutanol; ketoacid decarboxylase; metabolic engineering; microaerobic.
© 2019 The Authors. Engineering in Life Sciences published by WILEY‐VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

Similar articles
-
Corynebacterium glutamicum tailored for efficient isobutanol production.Appl Environ Microbiol. 2011 May;77(10):3300-10. doi: 10.1128/AEM.02972-10. Epub 2011 Mar 25. Appl Environ Microbiol. 2011. PMID: 21441331 Free PMC article.
-
Engineering Corynebacterium glutamicum for isobutanol production.Appl Microbiol Biotechnol. 2010 Jul;87(3):1045-55. doi: 10.1007/s00253-010-2522-6. Epub 2010 Apr 8. Appl Microbiol Biotechnol. 2010. PMID: 20376637 Free PMC article.
-
Comparative assessment of native and heterologous 2-oxo acid decarboxylases for application in isobutanol production by Saccharomyces cerevisiae.Biotechnol Biofuels. 2015 Dec 1;8:204. doi: 10.1186/s13068-015-0374-0. eCollection 2015. Biotechnol Biofuels. 2015. PMID: 26628917 Free PMC article.
-
Isobutanol production in engineered Saccharomyces cerevisiae by overexpression of 2-ketoisovalerate decarboxylase and valine biosynthetic enzymes.Bioprocess Biosyst Eng. 2012 Nov;35(9):1467-75. doi: 10.1007/s00449-012-0736-y. Epub 2012 Apr 28. Bioprocess Biosyst Eng. 2012. PMID: 22543927
-
Isobutanol and 2-ketoisovalerate production by Klebsiella pneumoniae via a native pathway.Metab Eng. 2017 Sep;43(Pt A):71-84. doi: 10.1016/j.ymben.2017.07.003. Epub 2017 Aug 9. Metab Eng. 2017. PMID: 28802880
Cited by
-
Microbial host engineering for sustainable isobutanol production from renewable resources.Appl Microbiol Biotechnol. 2024 Dec;108(1):33. doi: 10.1007/s00253-023-12821-9. Epub 2024 Jan 4. Appl Microbiol Biotechnol. 2024. PMID: 38175234 Review.
-
Identification and Characterization of a Novel Soluble Pyridine Nucleotide Transhydrogenase from Streptomyces avermitilis.Curr Microbiol. 2021 Dec 20;79(1):32. doi: 10.1007/s00284-021-02727-y. Curr Microbiol. 2021. PMID: 34931264
-
Development and immunological evaluation of an mRNA-based vaccine targeting Naegleria fowleri for the treatment of primary amoebic meningoencephalitis.Sci Rep. 2024 Jan 8;14(1):767. doi: 10.1038/s41598-023-51127-8. Sci Rep. 2024. PMID: 38191579 Free PMC article.
-
Cyperus esculentus var. sativus Adapts to Multiple Heavy Metal Stresses Through the Assembly of Endophytic Microbial Communities.Biology (Basel). 2025 Jan 16;14(1):83. doi: 10.3390/biology14010083. Biology (Basel). 2025. PMID: 39857313 Free PMC article.
-
Systems biology of electrogenic Pseudomonas putida - multi-omics insights and metabolic engineering for enhanced 2-ketogluconate production.Microb Cell Fact. 2024 Sep 11;23(1):246. doi: 10.1186/s12934-024-02509-8. Microb Cell Fact. 2024. PMID: 39261865 Free PMC article.
References
-
- Eikmanns, B.J. and Blombach, B. , Isobutanol, in: Bisaria, V. S., Kondo, A. (Eds.), Bioprocessing of Renewable Resources to Commodity Bioproducts, John Wiley & Sons, Inc., Weinheim: 2014, pp. 327–352.
-
- Chen, C.‐T. and Liao, J.C. , Frontiers in microbial 1‐butanol and isobutanol production. FEMS Microbiol. Lett. 2016, 363, fnw020. - PubMed
-
- Atsumi, S. , Hanai, T. , Liao, J.C. , Non‐fermentative pathways for synthesis of branched‐chain higher alcohols as biofuels. Nature 2008, 451, 86–89. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
