Neuropeptides Modulate Local Astrocytes to Regulate Adult Hippocampal Neural Stem Cells
- PMID: 32877641
- PMCID: PMC7606593
- DOI: 10.1016/j.neuron.2020.07.039
Neuropeptides Modulate Local Astrocytes to Regulate Adult Hippocampal Neural Stem Cells
Abstract
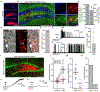
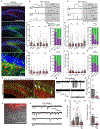
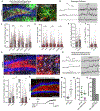
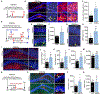
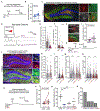
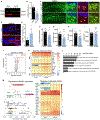
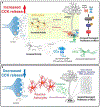
Neural stem cells (NSCs) in the dentate gyrus (DG) reside in a specialized local niche that supports their neurogenic proliferation to produce adult-born neurons throughout life. How local niche cells interact at the circuit level to ensure continuous neurogenesis from NSCs remains unknown. Here we report the role of endogenous neuropeptide cholecystokinin (CCK), released from dentate CCK interneurons, in regulating neurogenic niche cells and NSCs. Specifically, stimulating CCK release supports neurogenic proliferation of NSCs through a dominant astrocyte-mediated glutamatergic signaling cascade. In contrast, reducing dentate CCK induces reactive astrocytes, which correlates with decreased neurogenic proliferation of NSCs and upregulation of genes involved in immune processes. Our findings provide novel circuit-based information on how CCK acts on local astrocytes to regulate the key behavior of adult NSCs.
Keywords: adult neural stem cells; astrocytes; cholecystokinin; hippocampus; neurogenesis; neuroinflammation; neuropeptide.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

Comment in
-
GABAergic Interneurons Control Adult Neurogenesis but Astrocytes Have the Last Word.Neuron. 2020 Oct 28;108(2):226-228. doi: 10.1016/j.neuron.2020.09.041. Neuron. 2020. PMID: 33120019
References
-
- Araque A, Sanzgiri RP, Parpura V, and Haydon PG (1998). Calcium elevation in astrocytes causes an NMDA receptor-dependent increase in the frequency of miniature synaptic currents in cultured hippocampal neurons. The Journal of neuroscience : the official journal of the Society for Neuroscience 18, 6822–6829. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

