Most Commonly Mutated Genes in High-Grade Serous Ovarian Carcinoma Are Nonessential for Ovarian Surface Epithelial Stem Cell Transformation
- PMID: 32877668
- PMCID: PMC7545232
- DOI: 10.1016/j.celrep.2020.108086
Most Commonly Mutated Genes in High-Grade Serous Ovarian Carcinoma Are Nonessential for Ovarian Surface Epithelial Stem Cell Transformation
Abstract
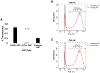
High-grade serous ovarian carcinoma (HGSOC) is the fifth leading cause of cancer-related deaths of women in the United States. Disease-associated mutations have been identified by the Cancer Genome Atlas Research Network. However, aside from mutations in TP53 or the RB1 pathway that are common in HGSOC, the contributions of mutation combinations are unclear. Here, we report CRISPR mutagenesis of 20 putative HGSOC driver genes to identify combinatorial disruptions of genes that transform either ovarian surface epithelium stem cells (OSE-SCs) or non-stem cells (OSE-NSs). Our results support the OSE-SC theory of HGSOC initiation and suggest that most commonly mutated genes in HGSOC have no effect on OSE-SC transformation initiation. Our results indicate that disruption of TP53 and PTEN, combined with RB1 disruption, constitutes a core set of mutations driving efficient transformation in vitro. The combined data may contribute to more accurate modeling of HGSOC development.
Keywords: CRISPR/Cas9; mutagenesis; ovarian cancer; stem cells; tumor drivers.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures
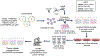


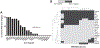

References
-
- Auersperg N (2013). Ovarian surface epithelium as a source of ovarian cancers: unwarranted speculation or evidence-based hypothesis? Gynecol. Oncol 130,246–251. - PubMed
-
- Auersperg N, Wong AS, Choi KC, Kang SK, and Leung PC (2001). Ovarian surface epithelium: biology, endocrinology, and pathology. Endocr. Rev 22, 255–288. - PubMed
-
- Auersperg N, Woo MMM, and Gilks CB (2008). The origin of ovarian carcinomas: a developmental view. Gynecol. Oncol 110, 452–454. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

