STING cyclic dinucleotide sensing originated in bacteria
- PMID: 32877915
- PMCID: PMC7572726
- DOI: 10.1038/s41586-020-2719-5
STING cyclic dinucleotide sensing originated in bacteria
Abstract
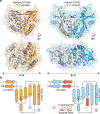
Stimulator of interferon genes (STING) is a receptor in human cells that senses foreign cyclic dinucleotides that are released during bacterial infection and in endogenous cyclic GMP-AMP signalling during viral infection and anti-tumour immunity1-5. STING shares no structural homology with other known signalling proteins6-9, which has limited attempts at functional analysis and prevented explanation of the origin of cyclic dinucleotide signalling in mammalian innate immunity. Here we reveal functional STING homologues encoded within prokaryotic defence islands, as well as a conserved mechanism of signal activation. Crystal structures of bacterial STING define a minimal homodimeric scaffold that selectively responds to cyclic di-GMP synthesized by a neighbouring cGAS/DncV-like nucleotidyltransferase (CD-NTase) enzyme. Bacterial STING domains couple the recognition of cyclic dinucleotides with the formation of protein filaments to drive oligomerization of TIR effector domains and rapid NAD+ cleavage. We reconstruct the evolutionary events that followed the acquisition of STING into metazoan innate immunity, and determine the structure of a full-length TIR-STING fusion from the Pacific oyster Crassostrea gigas. Comparative structural analysis demonstrates how metazoan-specific additions to the core STING scaffold enabled a switch from direct effector function to regulation of antiviral transcription. Together, our results explain the mechanism of STING-dependent signalling and reveal the conservation of a functional cGAS-STING pathway in prokaryotic defence against bacteriophages.
Conflict of interest statement
Figures











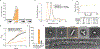

Comment in
-
Bacteria sting viral invaders.Nature. 2020 Oct;586(7829):363-364. doi: 10.1038/d41586-020-02712-8. Nature. 2020. PMID: 32989308 No abstract available.
References
-
- Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P & Shu HB The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity (2008) 29, 538–550 - PubMed
-
- Ablasser A & Chen ZJ cGAS in action: Expanding roles in immunity and inflammation. Science (2019) 363 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

