Circular RNAs: A New Piece in the Colorectal Cancer Puzzle
- PMID: 32878117
- PMCID: PMC7564116
- DOI: 10.3390/cancers12092464
Circular RNAs: A New Piece in the Colorectal Cancer Puzzle
Abstract
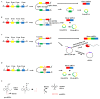
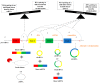
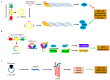
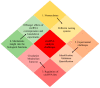
Colorectal cancer (CRC) is the third most fatal type of malignancy, worldwide. Despite the advances accomplished in the elucidation of its molecular base and the existing CRC biomarkers introduced in the clinical practice, additional research is required. Circular RNAs (circRNAs) constitute a new RNA type, formed by back-splicing of primary transcripts. They have been discovered during the 1970s but were characterized as by-products of aberrant splicing. However, the modern high-throughput approaches uncovered their widespread expression; therefore, several questions were raised regarding their potential biological roles. During the last years, great progress has been achieved in the elucidation of their functions: circRNAs can act as microRNA sponges, transcription regulators, and interfere with splicing, as well. Furthermore, they are heavily involved in various human pathological states, including cancer, and could serve as diagnostic and prognostic biomarkers in several diseases. Particularly in CRC, aberrant expression of circRNAs has been observed. More specifically, these molecules either inhibit or promote colorectal carcinogenesis by regulating different molecules and signaling pathways. The present review discusses the characteristics and functions of circRNA, prior to analyzing the multifaceted role of these molecules in CRC and their potential value as biomarkers and therapeutic targets.
Keywords: RNA splicing; circRNA; circularization; gastrointestinal cancer; microRNA sponges; peptide translation; regulation of carcinogenesis; therapeutic targets; transcription regulation; tumor biomarkers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources

