Molecular Changes in Tissue Proteome during Prostate Cancer Development: Proof-of-Principle Investigation
- PMID: 32878211
- PMCID: PMC7554904
- DOI: 10.3390/diagnostics10090655
Molecular Changes in Tissue Proteome during Prostate Cancer Development: Proof-of-Principle Investigation
Abstract
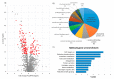
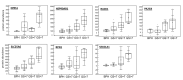
(1) Background: Prostate cancer (PCa) is characterized by high heterogeneity. The aim of this study was to investigate molecular alterations underlying PCa development based on proteomics data. (2) Methods: Liquid chromatography coupled to tandem mass spectrometry was conducted for 22 fresh-frozen tissue specimens from patients with benign prostatic hyperplasia (BPH, n = 5) and PCa (n = 17). Mann Whitney test was used to define significant differences between the two groups. Association of protein abundance with PCa progression was evaluated using Spearman correlation, followed by verification through investigating the Prostate Cancer Transcriptome Atlas. Functional enrichment and interactome analysis were carried out using Metascape and String. (3) Results: Proteomics analysis identified 1433 proteins, including 145 proteins as differentially abundant between patients with PCa and BPH. In silico analysis revealed alterations in several pathways and hallmarks implicated in metabolism and signalling, represented by 67 proteins. Among the latter, 21 proteins were correlated with PCa progression at both the protein and mRNA levels. Interactome analysis of these 21 proteins predicted interactions between Myc proto-oncogene (MYC) targets, protein processing in the endoplasmic reticulum, and oxidative phosphorylation, with MYC targets having a central role. (4) Conclusions: Tissue proteomics allowed for characterization of proteins and pathways consistently affected during PCa development. Further validation of these findings is required.
Keywords: drug target; personalized medicine; prostate cancer; proteomics; tissue.
Conflict of interest statement
H.M. is cofounder and co-owner of Mosaiques Diagnostics. A.L. and M.F. are employees of Mosaiques Diagnostics GmbH. The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures


References
-
- Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Global Cancer Observatory: Cancer Today. International Agency for Research on Cancer; Lyon, France: 2018. [(accessed on 14 July 2020)]. Available online: https://gco.Iarc.Fr/today.
-
- Ploussard G., Epstein J.I., Montironi R., Carroll P.R., Wirth M., Grimm M.O., Bjartell A.S., Montorsi F., Freedland S.J., Erbersdobler A., et al. The contemporary concept of significant versus insignificant prostate cancer. Eur. Urol. 2011;60:291–303. doi: 10.1016/j.eururo.2011.05.006. - DOI - PubMed
-
- Howlader N., Noone A.M., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. Seer Cancer Statistics Review, 1975–2017. National Cancer Institute; Bethesda, MD, USA: 2020.
-
- Moreira D.M., Howard L.E., Sourbeer K.N., Amarasekara H.S., Chow L.C., Cockrell D.C., Pratson C.L., Hanyok B.T., Aronson W.J., Kane C.J., et al. Predicting time from metastasis to overall survival in castration-resistant prostate cancer: Results from search. Clin. Genitourin. Cancer. 2017;15:60–66.e62. doi: 10.1016/j.clgc.2016.08.018. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

