Impact of lockdown on Covid-19 case fatality rate and viral mutations spread in 7 countries in Europe and North America
- PMID: 32878627
- PMCID: PMC7463225
- DOI: 10.1186/s12967-020-02501-x
Impact of lockdown on Covid-19 case fatality rate and viral mutations spread in 7 countries in Europe and North America
Abstract
Background: Severe acute respiratory syndrome CoV-2 (SARS-CoV-2) caused the first coronavirus disease 2019 (COVID-19) outbreak in China and has become a public health emergency of international concern. SARS-CoV-2 outbreak has been declared a pandemic by WHO on March 11th, 2020 and the same month several Countries put in place different lockdown restrictions and testing strategies in order to contain the spread of the virus.
Methods: The calculation of the Case Fatality Rate of SARS-CoV-2 in the Countries selected was made by using the data available at https://github.com/owid/covi-19-data/tree/master/public/data . Case fatality rate was calculated as the ratio between the death cases due to COVID-19, over the total number of SARS-CoV-2 reported cases 14 days before. Standard Case Fatality Rate values were normalized by the Country-specific ρ factor, i.e. the number of PCR tests/1 million inhabitants over the number of reported cases/1 million inhabitants. Case-fatality rates between Countries were compared using proportion test. Post-hoc analysis in the case of more than two groups was performed using pairwise comparison of proportions and p value was adjusted using Holm method. We also analyzed 487 genomic sequences from the GISAID database derived from patients infected by SARS-CoV-2 from January 2020 to April 2020 in Italy, Spain, Germany, France, Sweden, UK and USA. SARS-CoV-2 reference genome was obtained from the GenBank database (NC_045512.2). Genomes alignment was performed using Muscle and Jalview software. We, then, calculated the Case Fatality Rate of SARS-CoV-2 in the Countries selected.
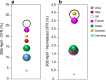
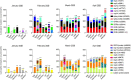
Results: In this study we analyse how different lockdown strategies and PCR testing capability adopted by Italy, France, Germany, Spain, Sweden, UK and USA have influenced the Case Fatality Rate and the viral mutations spread. We calculated case fatality rates by dividing the death number of a specific day by the number of patients with confirmed COVID-19 infection observed 14 days before and normalized by a ρ factor which takes into account the diagnostic PCR testing capability of each Country and the number of positive cases detected. We notice the stabilization of a clear pattern of mutations at sites nt241, nt3037, nt14408 and nt23403. A novel nonsynonymous SARS-CoV-2 mutation in the spike protein (nt24368) has been found in genomes sequenced in Sweden, which enacted a soft lockdown strategy.
Conclusions: Strict lockdown strategies together with a wide diagnostic PCR testing of the population were correlated with a relevant decline of the case fatality rate in different Countries. The emergence of specific patterns of mutations concomitant with the decline in case fatality rate needs further confirmation and their biological significance remains unclear.
Keywords: COVID-19; Case fatality rate; Europe; Lockdown strategy; Mutation; SARS-CoV-2; Testing capacity; US.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Comment in
-
Can reasoned mass testing impact covid-19 outcomes in wide community contexts? An evidence-based opinion.Pathog Glob Health. 2021 May;115(3):203-207. doi: 10.1080/20477724.2021.1878444. Epub 2021 Mar 9. Pathog Glob Health. 2021. PMID: 33686907 Free PMC article.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team Novel coronavirus from patients with pneumonia in China. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001017. - DOI - PMC - PubMed
-
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003 doi: 10.1056/NEJMoa030781. - DOI - PubMed
-
- Kuiken T, Fouchier RA, Schutten M, Rimmelzwaan GF, van Amerongen G, van Riel D, Laman JD, de Jong T, van Doornum G, Lim W, Ling AE, Chan PK, Tam JS, Zambon MC, Gopal R, Drosten C, van der Werf S, Escriou N, Manuguerra JC, Stöhr K, Osterhaus AD. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003 doi: 10.1016/S0140-6736(03)13967-0. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

