Retinoic Acid Inducible Gene I and Protein Kinase R, but Not Stress Granules, Mediate the Proinflammatory Response to Yellow Fever Virus
- PMID: 32878892
- PMCID: PMC7592215
- DOI: 10.1128/JVI.00403-20
Retinoic Acid Inducible Gene I and Protein Kinase R, but Not Stress Granules, Mediate the Proinflammatory Response to Yellow Fever Virus
Abstract
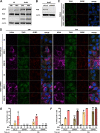
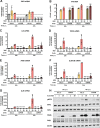
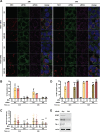
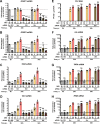
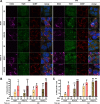
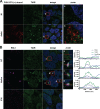
Yellow fever virus (YFV) is an RNA virus primarily targeting the liver. Severe YF cases are responsible for hemorrhagic fever, plausibly precipitated by excessive proinflammatory cytokine response. Pathogen recognition receptors (PRRs), such as the cytoplasmic retinoic acid inducible gene I (RIG-I)-like receptors (RLRs), and the viral RNA sensor protein kinase R (PKR), are known to initiate a proinflammatory response upon recognition of viral genomes. Here, we sought to reveal the main determinants responsible for the acute cytokine expression occurring in human hepatocytes following YFV infection. Using a RIG-I-defective human hepatoma cell line, we found that RIG-I largely contributes to cytokine secretion upon YFV infection. In infected RIG-I-proficient hepatoma cells, RIG-I was localized in stress granules. These granules are large aggregates of stalled translation preinitiation complexes known to concentrate RLRs and PKR and are so far recognized as hubs orchestrating RNA virus sensing. Stable knockdown of PKR in hepatoma cells revealed that PKR contributes to both stress granule formation and cytokine induction upon YFV infection. However, stress granule disruption did not affect the cytokine response to YFV infection, as assessed by small interfering RNA (siRNA)-knockdown-mediated inhibition of stress granule assembly. Finally, no viral RNA was detected in stress granules using a fluorescence in situ hybridization approach coupled with immunofluorescence. Our findings suggest that both RIG-I and PKR mediate proinflammatory cytokine induction in YFV-infected hepatocytes, in a stress granule-independent manner. Therefore, by showing the uncoupling of the cytokine response from the stress granule formation, our model challenges the current view in which stress granules are required for the mounting of the acute antiviral response.IMPORTANCE Yellow fever is a mosquito-borne acute hemorrhagic disease caused by yellow fever virus (YFV). The mechanisms responsible for its pathogenesis remain largely unknown, although increased inflammation has been linked to worsened outcome. YFV targets the liver, where it primarily infects hepatocytes. We found that two RNA-sensing proteins, RIG-I and PKR, participate in the induction of proinflammatory mediators in human hepatocytes infected with YFV. We show that YFV infection promotes the formation of cytoplasmic structures, termed stress granules, in a PKR- but not RIG-I-dependent manner. While stress granules were previously postulated to be essential platforms for immune activation, we found that they are not required for the production of proinflammatory mediators upon YFV infection. Collectively, our work uncovered molecular events triggered by the replication of YFV, which could prove instrumental in clarifying the pathogenesis of the disease, with possible repercussions for disease management.
Keywords: cytokines; flavivirus; innate immunity; interferons; liver inflammation; pattern recognition receptors; stress granules; yellow fever virus.
Copyright © 2020 American Society for Microbiology.
Figures
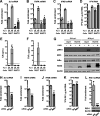
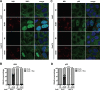
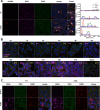
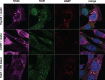
References
-
- Gubler DJ, Kuno G, Markoff L. 2007. Flaviviruses, p 1153–1252. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

