Pooled analysis of radiation hybrids identifies loci for growth and drug action in mammalian cells
- PMID: 32878976
- PMCID: PMC7605260
- DOI: 10.1101/gr.262204.120
Pooled analysis of radiation hybrids identifies loci for growth and drug action in mammalian cells
Abstract
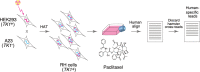
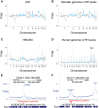
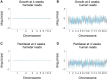
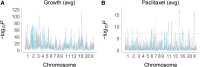
Genetic screens in mammalian cells commonly focus on loss-of-function approaches. To evaluate the phenotypic consequences of extra gene copies, we used bulk segregant analysis (BSA) of radiation hybrid (RH) cells. We constructed six pools of RH cells, each consisting of ∼2500 independent clones, and placed the pools under selection in media with or without paclitaxel. Low pass sequencing identified 859 growth loci, 38 paclitaxel loci, 62 interaction loci, and three loci for mitochondrial abundance at genome-wide significance. Resolution was measured as ∼30 kb, close to single-gene. Divergent properties were displayed by the RH-BSA growth genes compared to those from loss-of-function screens, refuting the balance hypothesis. In addition, enhanced retention of human centromeres in the RH pools suggests a new approach to functional dissection of these chromosomal elements. Pooled analysis of RH cells showed high power and resolution and should be a useful addition to the mammalian genetic toolkit.
© 2020 Khan et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Aoki J, Kai W, Kawabata Y, Ozaki A, Yoshida K, Tsuzaki T, Fuji K, Koyama T, Sakamoto T, Araki K. 2014. Construction of a radiation hybrid panel and the first yellowtail (Seriola quinqueradiata) radiation hybrid map using a nanofluidic dynamic array. BMC Genomics 15: 165 10.1186/1471-2164-15-165 - DOI - PMC - PubMed
