A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components
- PMID: 32878980
- PMCID: PMC8269000
- DOI: 10.1126/scitranslmed.aaz4589
A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components
Abstract
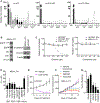
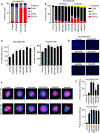
Acquired resistance to tyrosine kinase inhibitors (TKIs) of epidermal growth factor receptor (EGFR) remains a clinical challenge. Especially challenging are cases in which resistance emerges through EGFR-independent mechanisms, such as through pathways that promote epithelial-to-mesenchymal transition (EMT). Through an integrated transcriptomic, proteomic, and drug screening approach, we identified activation of the yes-associated protein (YAP) and forkhead box protein M1 (FOXM1) axis as a driver of EMT-associated EGFR TKI resistance. EGFR inhibitor resistance was associated with broad multidrug resistance that extended across multiple chemotherapeutic and targeted agents, consistent with the difficulty of effectively treating resistant disease. EGFR TKI-resistant cells displayed increased abundance of spindle assembly checkpoint (SAC) proteins, including polo-like kinase 1 (PLK1), Aurora kinases, survivin, and kinesin spindle protein (KSP). Moreover, EGFR TKI-resistant cells exhibited vulnerability to SAC inhibitors. Increased activation of the YAP/FOXM1 axis mediated an increase in the abundance of SAC components in resistant cells. The clinical relevance of these finding was indicated by evaluation of specimens from patients with EGFR mutant lung cancer, which showed that high FOXM1 expression correlated with expression of genes encoding SAC proteins and was associated with a worse clinical outcome. These data revealed the YAP/FOXM1 axis as a central regulator of EMT-associated EGFR TKI resistance and that this pathway, along with SAC components, are therapeutic vulnerabilities for targeting this multidrug-resistant phenotype.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures





References
-
- Mok TS et al., Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. The New England journal of medicine 361, 947–957 (2009). - PubMed
-
- Rosell R et al., Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The Lancet. Oncology 13, 239–246 (2012). - PubMed
-
- Maemondo M et al., Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. The New England journal of medicine 362, 2380–2388 (2010). - PubMed
-
- Sequist LV et al., First-line gefitinib in patients with advanced non-small-cell lung cancer harboring somatic EGFR mutations. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 26, 2442–2449 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

