Circadian control of brain glymphatic and lymphatic fluid flow
- PMID: 32879313
- PMCID: PMC7468152
- DOI: 10.1038/s41467-020-18115-2
Circadian control of brain glymphatic and lymphatic fluid flow
Abstract
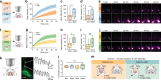
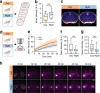
The glymphatic system is a network of perivascular spaces that promotes movement of cerebrospinal fluid (CSF) into the brain and clearance of metabolic waste. This fluid transport system is supported by the water channel aquaporin-4 (AQP4) localized to vascular endfeet of astrocytes. The glymphatic system is more effective during sleep, but whether sleep timing promotes glymphatic function remains unknown. We here show glymphatic influx and clearance exhibit endogenous, circadian rhythms peaking during the mid-rest phase of mice. Drainage of CSF from the cisterna magna to the lymph nodes exhibits daily variation opposite to glymphatic influx, suggesting distribution of CSF throughout the animal depends on time-of-day. The perivascular polarization of AQP4 is highest during the rest phase and loss of AQP4 eliminates the day-night difference in both glymphatic influx and drainage to the lymph nodes. We conclude that CSF distribution is under circadian control and that AQP4 supports this rhythm.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Kreutzmann JC, Havekes R, Abel T, Meerlo P. Sleep deprivation and hippocampal vulnerability: changes in neuronal plasticity, neurogenesis and cognitive function. Neuroscience. 2015;309:173–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

