The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer's disease
- PMID: 32879487
- PMCID: PMC7919141
- DOI: 10.1038/s41586-020-2681-2
The innate immunity protein IFITM3 modulates γ-secretase in Alzheimer's disease
Abstract
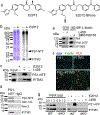
Innate immunity is associated with Alzheimer's disease1, but the influence of immune activation on the production of amyloid-β is unknown2,3. Here we identify interferon-induced transmembrane protein 3 (IFITM3) as a γ-secretase modulatory protein, and establish a mechanism by which inflammation affects the generation of amyloid-β. Inflammatory cytokines induce the expression of IFITM3 in neurons and astrocytes, which binds to γ-secretase and upregulates its activity, thereby increasing the production of amyloid-β. The expression of IFITM3 is increased with ageing and in mouse models that express familial Alzheimer's disease genes. Furthermore, knockout of IFITM3 reduces γ-secretase activity and the formation of amyloid plaques in a transgenic mouse model (5xFAD) of early amyloid deposition. IFITM3 protein is upregulated in tissue samples from a subset of patients with late-onset Alzheimer's disease that exhibit higher γ-secretase activity. The amount of IFITM3 in the γ-secretase complex has a strong and positive correlation with γ-secretase activity in samples from patients with late-onset Alzheimer's disease. These findings reveal a mechanism in which γ-secretase is modulated by neuroinflammation via IFITM3 and the risk of Alzheimer's disease is thereby increased.
Conflict of interest statement
COI
LYM is co-inventor of intellectual property (assay for gamma secretase activity and screening method for gamma secretase inhibitors) owned by MSKCC and licensed to Jiangsu Continental Medical Development.
Figures
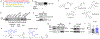
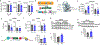


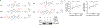


Comment in
-
Activity of Alzheimer's γ-secretase is linked to changes of interferon-induced transmembrane proteins (IFITM) in innate immunity.Mol Neurodegener. 2020 Nov 12;15(1):69. doi: 10.1186/s13024-020-00417-0. Mol Neurodegener. 2020. PMID: 33183335 Free PMC article.
References
References and Notes:
-
- De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active gamma-Secretase complex. Neuron 38, 9–12 (2003). - PubMed
Methods references
-
- Li YM et al. Photoactivated gamma-secretase inhibitors directed to the active site covalently label presenilin 1. Nature 405, 689–694 (2000). - PubMed
-
- Chun J, Yin YI, Yang G, Tarassishin L & Li YM Stereoselective Synthesis of Photoreactive Peptidomimetic gamma-Secretase Inhibitors. J Org Chem 69, 7344–7347 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AG026660/AG/NIA NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P30 CA008748/CA/NCI NIH HHS/United States
- R01 AG061350/AG/NIA NIH HHS/United States
- R01 AG018440/AG/NIA NIH HHS/United States
- R01 AG006170/AG/NIA NIH HHS/United States
- RF1 AG057593/AG/NIA NIH HHS/United States
- R01 NS096275/NS/NINDS NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01 AG057907/AG/NIA NIH HHS/United States
- S10 OD018522/OD/NIH HHS/United States
- S10 OD026880/OD/NIH HHS/United States
- K01 AG062683/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

