Cancer SLC43A2 alters T cell methionine metabolism and histone methylation
- PMID: 32879489
- PMCID: PMC7486248
- DOI: 10.1038/s41586-020-2682-1
Cancer SLC43A2 alters T cell methionine metabolism and histone methylation
Abstract
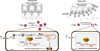
Abnormal epigenetic patterns correlate with effector T cell malfunction in tumours1-4, but the cause of this link is unknown. Here we show that tumour cells disrupt methionine metabolism in CD8+ T cells, thereby lowering intracellular levels of methionine and the methyl donor S-adenosylmethionine (SAM) and resulting in loss of dimethylation at lysine 79 of histone H3 (H3K79me2). Loss of H3K79me2 led to low expression of STAT5 and impaired T cell immunity. Mechanistically, tumour cells avidly consumed methionine and outcompeted T cells for methionine by expressing high levels of the methionine transporter SLC43A2. Genetic and biochemical inhibition of tumour SLC43A2 restored H3K79me2 in T cells, thereby boosting spontaneous and checkpoint-induced tumour immunity. Moreover, methionine supplementation improved the expression of H3K79me2 and STAT5 in T cells, and this was accompanied by increased T cell immunity in tumour-bearing mice and patients with colon cancer. Clinically, tumour SLC43A2 correlated negatively with T cell histone methylation and functional gene signatures. Our results identify a mechanistic connection between methionine metabolism, histone patterns, and T cell immunity in the tumour microenvironment. Thus, cancer methionine consumption is an immune evasion mechanism, and targeting cancer methionine signalling may provide an immunotherapeutic approach.
Conflict of interest statement
Declaration of Interests
The authors declare no competing interests.
Figures





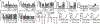

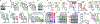

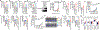
Comment in
-
Methio "mine"! Cancer cells steal methionine and impair CD8 T-cell function.Immunol Cell Biol. 2020 Sep;98(8):623-625. doi: 10.1111/imcb.12385. Epub 2020 Aug 25. Immunol Cell Biol. 2020. PMID: 32842169
-
A Tug-of-War Over Methionine.Cell Metab. 2020 Nov 3;32(5):699-701. doi: 10.1016/j.cmet.2020.10.016. Cell Metab. 2020. PMID: 33147482
References
Publication types
MeSH terms
Substances
Grants and funding
- CA099985/CA/NCI NIH HHS/United States
- R01 CA193136/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- CA217648/CA/NCI NIH HHS/United States
- CA152470/CA/NCI NIH HHS/United States
- R01 CA152470/CA/NCI NIH HHS/United States
- R01 CA123088/CA/NCI NIH HHS/United States
- U24 DK097153/DK/NIDDK NIH HHS/United States
- R37 CA237421/CA/NCI NIH HHS/United States
- R01 CA099985/CA/NCI NIH HHS/United States
- R01 CA217510/CA/NCI NIH HHS/United States
- DK097153/NH/NIH HHS/United States
- R01 CA190176/CA/NCI NIH HHS/United States
- R01 CA214911/CA/NCI NIH HHS/United States
- CA123088/CA/NCI NIH HHS/United States
- CA193136/CA/NCI NIH HHS/United States
- T32 CA009676/CA/NCI NIH HHS/United States
- R01 CA248430/CA/NCI NIH HHS/United States
- R01 CA211016/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

