Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis
- PMID: 32881682
- PMCID: PMC10339462
- DOI: 10.1109/TMI.2020.3021387
Pathomic Fusion: An Integrated Framework for Fusing Histopathology and Genomic Features for Cancer Diagnosis and Prognosis
Abstract
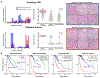
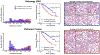
Cancer diagnosis, prognosis, mymargin and therapeutic response predictions are based on morphological information from histology slides and molecular profiles from genomic data. However, most deep learning-based objective outcome prediction and grading paradigms are based on histology or genomics alone and do not make use of the complementary information in an intuitive manner. In this work, we propose Pathomic Fusion, an interpretable strategy for end-to-end multimodal fusion of histology image and genomic (mutations, CNV, RNA-Seq) features for survival outcome prediction. Our approach models pairwise feature interactions across modalities by taking the Kronecker product of unimodal feature representations, and controls the expressiveness of each representation via a gating-based attention mechanism. Following supervised learning, we are able to interpret and saliently localize features across each modality, and understand how feature importance shifts when conditioning on multimodal input. We validate our approach using glioma and clear cell renal cell carcinoma datasets from the Cancer Genome Atlas (TCGA), which contains paired whole-slide image, genotype, and transcriptome data with ground truth survival and histologic grade labels. In a 15-fold cross-validation, our results demonstrate that the proposed multimodal fusion paradigm improves prognostic determinations from ground truth grading and molecular subtyping, as well as unimodal deep networks trained on histology and genomic data alone. The proposed method establishes insight and theory on how to train deep networks on multimodal biomedical data in an intuitive manner, which will be useful for other problems in medicine that seek to combine heterogeneous data streams for understanding diseases and predicting response and resistance to treatment. Code and trained models are made available at: https://github.com/mahmoodlab/PathomicFusion.
Figures

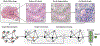


References
-
- Wen PY and Kesari S, “Malignant gliomas in adults,” New England Journal of Medicine, vol. 359, no. 5, pp. 492–507, Jul. 2008. - PubMed
-
- Nayak L, Lee EQ, and Wen PY, “Epidemiology of brain metastases,” Current Oncology Reports, vol. 14, no. 1, pp. 48–54, Oct. 2011. - PubMed
-
- Aldape K, Zadeh G, Mansouri S, Reifenberger G, and von Deimling A, “Glioblastoma: pathology, molecular mechanisms and markers,” Acta Neuropathologica, vol. 129, no. 6, pp. 829–848, May 2015. - PubMed
-
- Shanes J, Ghali J, Billingham M, Ferrans V, Fenoglio J, Edwards W, Tsai C, Saffitz J, Isner J, and Furner S, “Interobserver variability in the pathologic interpretation of endomyocardial biopsy results.” Circulation, vol. 75, no. 2, pp. 401–405, 1987. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

