Phosphorylation controls RNA binding and transcription by the influenza virus polymerase
- PMID: 32881973
- PMCID: PMC7494117
- DOI: 10.1371/journal.ppat.1008841
Phosphorylation controls RNA binding and transcription by the influenza virus polymerase
Abstract
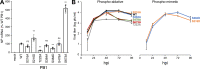
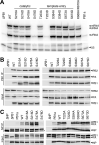
The influenza virus polymerase transcribes and replicates the viral genome. The proper timing and balance of polymerase activity is important for successful replication. Genome replication is controlled in part by phosphorylation of NP that regulates assembly of the replication machinery. However, it remains unclear whether phosphorylation directly regulated polymerase activity. Here we identified polymerase phosphosites that control its function. Mutating phosphosites in the catalytic subunit PB1 altered polymerase activity and virus replication. Biochemical analyses revealed phosphorylation events that disrupted global polymerase function by blocking the NTP entry channel or preventing RNA binding. We also identified a regulatory site that split polymerase function by specifically suppressing transcription. These experiments show that host kinases phospho-regulate viral RNA synthesis directly by modulating polymerase activity and indirectly by controlling assembly of replication machinery. Further, they suggest polymerase phosphorylation may bias replication versus transcription at discrete times or locations during the infectious cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Richard C-A, Rincheval V, Lassoued S, Fix J, Cardone C, Esneau C, et al. RSV hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. Schnell MJ, editor. PLOS Pathog. Public Library of Science; 2018;14: e1006920 10.1371/journal.ppat.1006920 - DOI - PMC - PubMed
-
- Chen L, Zhang S, Banerjee AK, Chen M. N-terminal phosphorylation of phosphoprotein of vesicular stomatitis virus is required for preventing nucleoprotein from binding to cellular RNAs and for functional template formation. J Virol. United States; 2013;87: 3177–3186. 10.1128/JVI.02761-12 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

