TBDB: a database of structurally annotated T-box riboswitch:tRNA pairs
- PMID: 32882008
- PMCID: PMC7778990
- DOI: 10.1093/nar/gkaa721
TBDB: a database of structurally annotated T-box riboswitch:tRNA pairs
Abstract
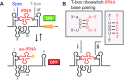
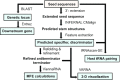
T-box riboswitches constitute a large family of tRNA-binding leader sequences that play a central role in gene regulation in many gram-positive bacteria. Accurate inference of the tRNA binding to T-box riboswitches is critical to predict their cis-regulatory activity. However, there is no central repository of information on the tRNA binding specificities of T-box riboswitches, and de novo prediction of binding specificities requires advanced knowledge of computational tools to annotate riboswitch secondary structure features. Here, we present the T-box Riboswitch Annotation Database (TBDB, https://tbdb.io), an open-access database with a collection of 23,535 T-box riboswitch sequences, spanning the major phyla of 3,632 bacterial species. Among structural predictions, the TBDB also identifies specifier sequences, cognate tRNA binding partners, and downstream regulatory targets. To our knowledge, the TBDB presents the largest collection of feature, sequence, and structural annotations carried out on this important family of regulatory RNA.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
New tRNA contacts facilitate ligand binding in a Mycobacterium smegmatis T box riboswitch.Proc Natl Acad Sci U S A. 2018 Apr 10;115(15):3894-3899. doi: 10.1073/pnas.1721254115. Epub 2018 Mar 26. Proc Natl Acad Sci U S A. 2018. PMID: 29581302 Free PMC article.
-
Hierarchical mechanism of amino acid sensing by the T-box riboswitch.Nat Commun. 2018 May 14;9(1):1896. doi: 10.1038/s41467-018-04305-6. Nat Commun. 2018. PMID: 29760498 Free PMC article.
-
T box riboswitches in Actinobacteria: translational regulation via novel tRNA interactions.Proc Natl Acad Sci U S A. 2015 Jan 27;112(4):1113-8. doi: 10.1073/pnas.1424175112. Epub 2015 Jan 12. Proc Natl Acad Sci U S A. 2015. PMID: 25583497 Free PMC article.
-
Sequence, structure, and stacking: specifics of tRNA anchoring to the T box riboswitch.RNA Biol. 2013 Dec;10(12):1761-4. doi: 10.4161/rna.26996. Epub 2013 Nov 4. RNA Biol. 2013. PMID: 24356646 Free PMC article. Review.
-
An evolving tale of two interacting RNAs-themes and variations of the T-box riboswitch mechanism.IUBMB Life. 2019 Aug;71(8):1167-1180. doi: 10.1002/iub.2098. Epub 2019 Jun 17. IUBMB Life. 2019. PMID: 31206978 Free PMC article. Review.
Cited by
-
Direct observation of tRNA-chaperoned folding of a dynamic mRNA ensemble.Nat Commun. 2023 Sep 6;14(1):5438. doi: 10.1038/s41467-023-41155-3. Nat Commun. 2023. PMID: 37673863 Free PMC article.
-
An ontology-based knowledge graph for representing interactions involving RNA molecules.Sci Data. 2024 Aug 22;11(1):906. doi: 10.1038/s41597-024-03673-7. Sci Data. 2024. PMID: 39174566 Free PMC article.
-
Cooperativity and Interdependency between RNA Structure and RNA-RNA Interactions.Noncoding RNA. 2021 Dec 15;7(4):81. doi: 10.3390/ncrna7040081. Noncoding RNA. 2021. PMID: 34940761 Free PMC article. Review.
-
Recognition of the tRNA structure: Everything everywhere but not all at once.Cell Chem Biol. 2024 Jan 18;31(1):36-52. doi: 10.1016/j.chembiol.2023.12.008. Epub 2023 Dec 29. Cell Chem Biol. 2024. PMID: 38159570 Free PMC article. Review.
-
The 2021 Nucleic Acids Research database issue and the online molecular biology database collection.Nucleic Acids Res. 2021 Jan 8;49(D1):D1-D9. doi: 10.1093/nar/gkaa1216. Nucleic Acids Res. 2021. PMID: 33396976 Free PMC article.
References
-
- Winkler W. Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Curr. Opin. Chem. Biol. 2005; 9:594–602. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

