Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2
- PMID: 32883050
- PMCID: PMC7552048
- DOI: 10.3390/v12090972
Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2
Abstract
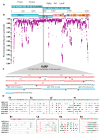
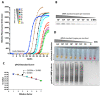
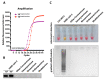
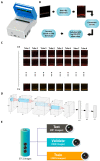
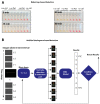
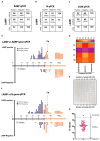
Until vaccines and effective therapeutics become available, the practical solution to transit safely out of the current coronavirus disease 19 (CoVID-19) lockdown may include the implementation of an effective testing, tracing and tracking system. However, this requires a reliable and clinically validated diagnostic platform for the sensitive and specific identification of SARS-CoV-2. Here, we report on the development of a de novo, high-resolution and comparative genomics guided reverse-transcribed loop-mediated isothermal amplification (LAMP) assay. To further enhance the assay performance and to remove any subjectivity associated with operator interpretation of results, we engineered a novel hand-held smart diagnostic device. The robust diagnostic device was further furnished with automated image acquisition and processing algorithms and the collated data was processed through artificial intelligence (AI) pipelines to further reduce the assay run time and the subjectivity of the colorimetric LAMP detection. This advanced AI algorithm-implemented LAMP (ai-LAMP) assay, targeting the RNA-dependent RNA polymerase gene, showed high analytical sensitivity and specificity for SARS-CoV-2. A total of ~200 coronavirus disease (CoVID-19)-suspected NHS patient samples were tested using the platform and it was shown to be reliable, highly specific and significantly more sensitive than the current gold standard qRT-PCR. Therefore, this system could provide an efficient and cost-effective platform to detect SARS-CoV-2 in resource-limited laboratories.
Keywords: LAMP; SARS-CoV-2; artificial intelligence; diagnosis; point of care.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. - DOI - PMC - PubMed
-
- Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

