Assessing Lysosomal Disorders in the NGS Era: Identification of Novel Rare Variants
- PMID: 32883051
- PMCID: PMC7503609
- DOI: 10.3390/ijms21176355
Assessing Lysosomal Disorders in the NGS Era: Identification of Novel Rare Variants
Abstract
Lysosomal storage diseases (LSDs) are a heterogeneous group of genetic disorders with variable degrees of severity and a broad phenotypic spectrum, which may overlap with a number of other conditions. While individually rare, as a group LSDs affect a significant number of patients, placing an important burden on affected individuals and their families but also on national health care systems worldwide. Here, we present our results on the use of an in-house customized next-generation sequencing (NGS) panel of genes related to lysosome function as a first-line molecular test for the diagnosis of LSDs. Ultimately, our goal is to provide a fast and effective tool to screen for virtually all LSDs in a single run, thus contributing to decrease the diagnostic odyssey, accelerating the time to diagnosis. Our study enrolled a group of 23 patients with variable degrees of clinical and/or biochemical suspicion of LSD. Briefly, NGS analysis data workflow, followed by segregation analysis allowed the characterization of approximately 41% of the analyzed patients and the identification of 10 different pathogenic variants, underlying nine LSDs. Importantly, four of those variants were novel, and, when applicable, their effect over protein structure was evaluated through in silico analysis. One of the novel pathogenic variants was identified in the GM2A gene, which is associated with an ultra-rare (or misdiagnosed) LSD, the AB variant of GM2 Gangliosidosis. Overall, this case series highlights not only the major advantages of NGS-based diagnostic approaches but also, to some extent, its limitations ultimately promoting a reflection on the role of targeted panels as a primary tool for the prompt characterization of LSD patients.
Keywords: CLN7; GM2 Gangliosidosis; GM2A gene; bioinformatics analysis; diagnostics odyssey; lysosomal storage diseases (LSDs); molecular genetic testing (MGT); next-generation sequencing (NGS).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


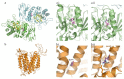

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

