Infarct Collagen Topography Regulates Fibroblast Fate via p38-Yes-Associated Protein Transcriptional Enhanced Associate Domain Signals
- PMID: 32883176
- PMCID: PMC7581544
- DOI: 10.1161/CIRCRESAHA.119.316162
Infarct Collagen Topography Regulates Fibroblast Fate via p38-Yes-Associated Protein Transcriptional Enhanced Associate Domain Signals
Abstract
Rationale: Myocardial infarction causes spatial variation in collagen organization and phenotypic diversity in fibroblasts, which regulate the heart's ECM (extracellular matrix). The relationship between collagen structure and fibroblast phenotype is poorly understood but could provide insights regarding the mechanistic basis for myofibroblast heterogeneity in the injured heart.
Objective: To investigate the role of collagen organization in cardiac fibroblast fate determination.
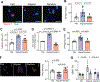
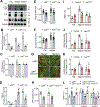
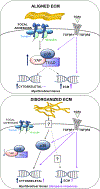
Methods and results: Biomimetic topographies were nanofabricated to recapitulate differential collagen organization in the infarcted mouse heart. Here, adult cardiac fibroblasts were freshly isolated and cultured on ECM topographical mimetics for 72 hours. Aligned mimetics caused cardiac fibroblasts to elongate while randomly organized topographies induced circular morphology similar to the disparate myofibroblast morphologies measured in vivo. Alignment cues also induced myofibroblast differentiation, as >60% of fibroblasts formed αSMA (α-smooth muscle actin) stress fibers and expressed myofibroblast-specific ECM genes like Postn (periostin). By contrast, random organization caused 38% of cardiac fibroblasts to express αSMA albeit with downregulated myofibroblast-specific ECM genes. Coupling topographical cues with the profibrotic agonist, TGFβ (transforming growth factor beta), additively upregulated myofibroblast-specific ECM genes independent of topography, but only fibroblasts on flat and randomly oriented mimetics had increased percentages of fibroblasts with αSMA stress fibers. Increased tension sensation at focal adhesions induced myofibroblast differentiation on aligned mimetics. These signals were transduced by p38-YAP (yes-associated protein)-TEAD (transcriptional enhanced associate domain) interactions, in which both p38 and YAP-TEAD (yes-associated protein transcriptional enhanced associate domain) binding were required for myofibroblast differentiation. By contrast, randomly oriented mimetics did not change focal adhesion tension sensation or enrich for p38-YAP-TEAD interactions, which explains the topography-dependent diversity in fibroblast phenotypes observed here.
Conclusions: Spatial variations in collagen organization regulate cardiac fibroblast phenotype through mechanical activation of p38-YAP-TEAD signaling, which likely contribute to myofibroblast heterogeneity in the infarcted myocardium.
Keywords: extracellular matrix; fibroblasts; fibrosis; myocardial infarction; nanotechnology.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

