A blueprint for academic laboratories to produce SARS-CoV-2 quantitative RT-PCR test kits
- PMID: 32883809
- PMCID: PMC7667971
- DOI: 10.1074/jbc.RA120.015434
A blueprint for academic laboratories to produce SARS-CoV-2 quantitative RT-PCR test kits
Abstract
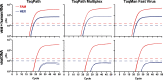
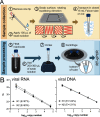
Widespread testing for the presence of the novel coronavirus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in individuals remains vital for controlling the COVID-19 pandemic prior to the advent of an effective treatment. Challenges in testing can be traced to an initial shortage of supplies, expertise, and/or instrumentation necessary to detect the virus by quantitative RT-PCR (RT-qPCR), the most robust, sensitive, and specific assay currently available. Here we show that academic biochemistry and molecular biology laboratories equipped with appropriate expertise and infrastructure can replicate commercially available SARS-CoV-2 RT-qPCR test kits and backfill pipeline shortages. The Georgia Tech COVID-19 Test Kit Support Group, composed of faculty, staff, and trainees across the biotechnology quad at Georgia Institute of Technology, synthesized multiplexed primers and probes and formulated a master mix composed of enzymes and proteins produced in-house. Our in-house kit compares favorably with a commercial product used for diagnostic testing. We also developed an environmental testing protocol to readily monitor surfaces for the presence of SARS-CoV-2. Our blueprint should be readily reproducible by research teams at other institutions, and our protocols may be modified and adapted to enable SARS-CoV-2 detection in more resource-limited settings.
Keywords: DNA polymerase; RNA; RT-qPCR; SARS-CoV-2; coronavirus; formulation; infectious disease; polymerase chain reaction (PCR); protein purification; reverse transcriptase; reverse transcription; ribonuclease inhibitor; virus.
© 2020 Mascuch et al.
Conflict of interest statement
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
Figures


Update of
-
A blueprint for academic labs to produce SARS-CoV-2 RT-qPCR test kits.medRxiv [Preprint]. 2020 Sep 1:2020.07.29.20163949. doi: 10.1101/2020.07.29.20163949. medRxiv. 2020. Update in: J Biol Chem. 2020 Nov 13;295(46):15438-15453. doi: 10.1074/jbc.RA120.015434. PMID: 32766604 Free PMC article. Updated. Preprint.
Similar articles
-
A blueprint for academic labs to produce SARS-CoV-2 RT-qPCR test kits.medRxiv [Preprint]. 2020 Sep 1:2020.07.29.20163949. doi: 10.1101/2020.07.29.20163949. medRxiv. 2020. Update in: J Biol Chem. 2020 Nov 13;295(46):15438-15453. doi: 10.1074/jbc.RA120.015434. PMID: 32766604 Free PMC article. Updated. Preprint.
-
Comparative evaluation of six nucleic acid amplification kits for SARS-CoV-2 RNA detection.Ann Clin Microbiol Antimicrob. 2021 May 22;20(1):38. doi: 10.1186/s12941-021-00443-w. Ann Clin Microbiol Antimicrob. 2021. PMID: 34022903 Free PMC article.
-
Simple, Inexpensive RNA Isolation and One-Step RT-qPCR Methods for SARS-CoV-2 Detection and General Use.Curr Protoc. 2021 Apr;1(4):e130. doi: 10.1002/cpz1.130. Curr Protoc. 2021. PMID: 33905620 Free PMC article.
-
RT-qPCR Testing and Performance Metrics in the COVID-19 Era.Int J Mol Sci. 2024 Aug 28;25(17):9326. doi: 10.3390/ijms25179326. Int J Mol Sci. 2024. PMID: 39273275 Free PMC article. Review.
-
An Update on Molecular Diagnostics for COVID-19.Front Cell Infect Microbiol. 2020 Nov 10;10:560616. doi: 10.3389/fcimb.2020.560616. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33244462 Free PMC article. Review.
Cited by
-
The concentration of single-stranded DNA-binding proteins is a critical factor in recombinase polymerase amplification (RPA), as revealed by insights from an open-source system.PeerJ. 2025 Aug 13;13:e19758. doi: 10.7717/peerj.19758. eCollection 2025. PeerJ. 2025. PMID: 40821985 Free PMC article.
-
Characteristics of patients with Coronavirus Disease 2019 (COVID-19) and seasonal influenza at time of hospital admission: a single center comparative study.BMC Infect Dis. 2021 Mar 17;21(1):271. doi: 10.1186/s12879-021-05957-4. BMC Infect Dis. 2021. PMID: 33731019 Free PMC article.
-
SARS-CoV-2 E Gene Variant Alters Analytical Sensitivity Characteristics of Viral Detection Using a Commercial Reverse Transcription-PCR Assay.J Clin Microbiol. 2021 Jun 18;59(7):e0007521. doi: 10.1128/JCM.00075-21. Epub 2021 Jun 18. J Clin Microbiol. 2021. PMID: 33903167 Free PMC article.
-
Analytical sensitivity and clinical performance of "COVID-19 RT-PCR Real TM FAST (CY5) (ATGen, Uruguay) and "ECUGEN SARS-CoV-2 RT-qPCR" (UDLA-STARNEWCORP, Ecuador)": High quality-low cost local SARS-CoV-2 tests for South America.PLoS Negl Trop Dis. 2022 Apr 13;16(4):e0010082. doi: 10.1371/journal.pntd.0010082. eCollection 2022 Apr. PLoS Negl Trop Dis. 2022. PMID: 35417456 Free PMC article.
-
Homebrew reagents for low cost RT-LAMP.medRxiv [Preprint]. 2021 May 12:2021.05.08.21256891. doi: 10.1101/2021.05.08.21256891. medRxiv. 2021. Update in: J Biomol Tech. 2021 Sep;32(3):114-120. doi: 10.7171/jbt.21-3203-006. PMID: 34013302 Free PMC article. Updated. Preprint.
References
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., et al. (2020) A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 10.1038/s41586-020-2008-3 - DOI - PMC - PubMed
-
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., et al. (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 10.1038/s41586-020-2012-7 - DOI - PMC - PubMed
-
- Patel R., Babady E., Theel E. S., Storch G. A., Pinsky B. A., George K. S., Smith T. C., and Bertuzzi S. (2020) Report from the American Society for Microbiology COVID-19 international summit, 23 March 2020: value of diagnostic testing for SARS–CoV-2/COVID-19. mBio 11, e00722–20 10.1128/mBio.00722-20 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

