In silico analysis predicting effects of deleterious SNPs of human RASSF5 gene on its structure and functions
- PMID: 32884013
- PMCID: PMC7471297
- DOI: 10.1038/s41598-020-71457-1
In silico analysis predicting effects of deleterious SNPs of human RASSF5 gene on its structure and functions
Abstract
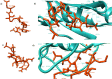
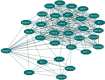
Ras association domain-containing protein 5 (RASSF5), one of the prospective biomarkers for tumors, generally plays a crucial role as a tumor suppressor. As deleterious effects can result from functional differences through SNPs, we sought to analyze the most deleterious SNPs of RASSF5 as well as predict the structural changes associated with the mutants that hamper the normal protein-protein interactions. We adopted both sequence and structure based approaches to analyze the SNPs of RASSF5 protein. We also analyzed the putative post translational modification sites as well as the altered protein-protein interactions that encompass various cascades of signals. Out of all the SNPs obtained from the NCBI database, only 25 were considered as highly deleterious by six in silico SNP prediction tools. Among them, upon analyzing the effect of these nsSNPs on the stability of the protein, we found 17 SNPs that decrease the stability. Significant deviation in the energy minimization score was observed in P350R, F321L, and R277W. Besides this, docking analysis confirmed that P350R, A319V, F321L, and R277W reduce the binding affinity of the protein with H-Ras, where P350R shows the most remarkable deviation. Protein-protein interaction analysis revealed that RASSF5 acts as a hub connecting two clusters consisting of 18 proteins and alteration in the RASSF5 may lead to disassociation of several signal cascades. Thus, based on these analyses, our study suggests that the reported functional SNPs may serve as potential targets for different proteomic studies, diagnosis and therapeutic interventions.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Comprehensive Characterization of the Coding and Non-Coding Single Nucleotide Polymorphisms in the Tumor Protein p63 (TP63) Gene Using In Silico Tools.Biomolecules. 2021 Nov 20;11(11):1733. doi: 10.3390/biom11111733. Biomolecules. 2021. PMID: 34827731 Free PMC article.
-
Engineered variants of the Ras effector protein RASSF5 (NORE1A) promote anticancer activities in lung adenocarcinoma.J Biol Chem. 2021 Dec;297(6):101353. doi: 10.1016/j.jbc.2021.101353. Epub 2021 Oct 27. J Biol Chem. 2021. PMID: 34717958 Free PMC article.
-
Predicting Deleterious Non-Synonymous Single Nucleotide Polymorphisms (nsSNPs) of HRAS Gene and In Silico Evaluation of Their Structural and Functional Consequences towards Diagnosis and Prognosis of Cancer.Biology (Basel). 2022 Nov 2;11(11):1604. doi: 10.3390/biology11111604. Biology (Basel). 2022. PMID: 36358305 Free PMC article.
-
Predicting the functional and structural consequences of nsSNPs in human methionine synthase gene using computational tools.Syst Biol Reprod Med. 2019 Aug;65(4):288-300. doi: 10.1080/19396368.2019.1568611. Epub 2019 Jan 24. Syst Biol Reprod Med. 2019. PMID: 30676783
-
The Emerging Roles of RASSF5 in Human Malignancy.Anticancer Agents Med Chem. 2018;18(3):314-322. doi: 10.2174/1871520617666170327120747. Anticancer Agents Med Chem. 2018. PMID: 28356010 Review.
Cited by
-
HBD-2 variants and SARS-CoV-2: New insights into inter-individual susceptibility.Front Immunol. 2022 Dec 9;13:1008463. doi: 10.3389/fimmu.2022.1008463. eCollection 2022. Front Immunol. 2022. PMID: 36569842 Free PMC article.
-
Comprehensive Characterization of the Coding and Non-Coding Single Nucleotide Polymorphisms in the Tumor Protein p63 (TP63) Gene Using In Silico Tools.Biomolecules. 2021 Nov 20;11(11):1733. doi: 10.3390/biom11111733. Biomolecules. 2021. PMID: 34827731 Free PMC article.
-
In silico evaluation of missense SNPs in cancer-associated Cystatin A protein and their potential to disrupt Cathepsin B interaction.Heliyon. 2025 Feb 5;11(3):e42478. doi: 10.1016/j.heliyon.2025.e42478. eCollection 2025 Feb 15. Heliyon. 2025. PMID: 40007784 Free PMC article.
-
Computational Analysis of Deleterious SNPs in NRAS to Assess Their Potential Correlation With Carcinogenesis.Front Genet. 2022 Aug 16;13:872845. doi: 10.3389/fgene.2022.872845. eCollection 2022. Front Genet. 2022. PMID: 36051694 Free PMC article.
-
Exploring the influence of pressure-induced semiconductor-to-metal transition on the physical properties of cubic perovskites FrXCl3 (X = Ge and Sn).Heliyon. 2024 Mar 17;10(7):e27581. doi: 10.1016/j.heliyon.2024.e27581. eCollection 2024 Apr 15. Heliyon. 2024. PMID: 38576570 Free PMC article.
References
-
- Vavvas D, Li X, Avruch J, Zhang X-F. Identification of Nore1 as a potential Ras effector. J. Biol. Chem. 1998;273:5439–5442. - PubMed
-
- Kumari G, Singhal PK, Suryaraja R, Mahalingam S. Functional interaction of the Ras effector RASSF5 with the tyrosine kinase Lck: critical role in nucleocytoplasmic transport and cell cycle regulation. J. Mol. Biol. 2010;397:89–109. - PubMed
-
- Li S, Teng J, Li H, Chen F, Zheng J. The emerging roles of RASSF5 in human malignancy. Anti-Cancer Agents Med. Chem. 2018;18:314–322. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

