Transcriptomic analysis of a Clostridium thermocellum strain engineered to utilize xylose: responses to xylose versus cellobiose feeding
- PMID: 32884054
- PMCID: PMC7471329
- DOI: 10.1038/s41598-020-71428-6
Transcriptomic analysis of a Clostridium thermocellum strain engineered to utilize xylose: responses to xylose versus cellobiose feeding
Abstract
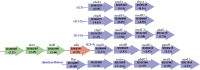
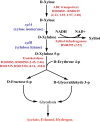
Clostridium (Ruminiclostridium) thermocellum is recognized for its ability to ferment cellulosic biomass directly, but it cannot naturally grow on xylose. Recently, C. thermocellum (KJC335) was engineered to utilize xylose through expressing a heterologous xylose catabolizing pathway. Here, we compared KJC335's transcriptomic responses to xylose versus cellobiose as the primary carbon source and assessed how the bacteria adapted to utilize xylose. Our analyses revealed 417 differentially expressed genes (DEGs) with log2 fold change (FC) >|1| and 106 highly DEGs (log2 FC >|2|). Among the DEGs, two putative sugar transporters, cbpC and cbpD, were up-regulated, suggesting their contribution to xylose transport and assimilation. Moreover, the up-regulation of specific transketolase genes (tktAB) suggests the importance of this enzyme for xylose metabolism. Results also showed remarkable up-regulation of chemotaxis and motility associated genes responding to xylose feeding, as well as widely varying gene expression in those encoding cellulosomal enzymes. For the down-regulated genes, several were categorized in gene ontology terms oxidation-reduction processes, ATP binding and ATPase activity, and integral components of the membrane. This study informs potentially critical, enabling mechanisms to realize the conceptually attractive Next-Generation Consolidated BioProcessing approach where a single species is sufficient for the co-fermentation of cellulose and hemicellulose.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Engineering cellulolytic bacterium Clostridium thermocellum to co-ferment cellulose- and hemicellulose-derived sugars simultaneously.Biotechnol Bioeng. 2018 Jul;115(7):1755-1763. doi: 10.1002/bit.26590. Epub 2018 Apr 10. Biotechnol Bioeng. 2018. PMID: 29537062
-
Engineering the cellulolytic bacterium, Clostridium thermocellum, to co-utilize hemicellulose.Metab Eng. 2024 May;83:193-205. doi: 10.1016/j.ymben.2024.03.008. Epub 2024 Apr 15. Metab Eng. 2024. PMID: 38631458
-
Transcriptomic and proteomic analyses of core metabolism in Clostridium termitidis CT1112 during growth on α-cellulose, xylan, cellobiose and xylose.BMC Microbiol. 2016 May 23;16:91. doi: 10.1186/s12866-016-0711-x. BMC Microbiol. 2016. PMID: 27215540 Free PMC article.
-
LacI Transcriptional Regulatory Networks in Clostridium thermocellum DSM1313.Appl Environ Microbiol. 2017 Feb 15;83(5):e02751-16. doi: 10.1128/AEM.02751-16. Print 2017 Mar 1. Appl Environ Microbiol. 2017. PMID: 28003194 Free PMC article.
-
[Mics of the Clostridium thermocellum in lignocellulose degradation--a review].Wei Sheng Wu Xue Bao. 2014 Feb 4;54(2):121-8. Wei Sheng Wu Xue Bao. 2014. PMID: 24818461 Review. Chinese.
Cited by
-
Thermophilic site-specific recombination system for rapid insertion of heterologous DNA into the Clostridium thermocellum chromosome.J Ind Microbiol Biotechnol. 2024 Dec 31;52:kuaf023. doi: 10.1093/jimb/kuaf023. J Ind Microbiol Biotechnol. 2024. PMID: 40844421 Free PMC article.
-
Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution.Microorganisms. 2021 Jul 4;9(7):1445. doi: 10.3390/microorganisms9071445. Microorganisms. 2021. PMID: 34361881 Free PMC article.
-
Genome-Wide Transcription Factor DNA Binding Sites and Gene Regulatory Networks in Clostridium thermocellum.Front Microbiol. 2021 Sep 7;12:695517. doi: 10.3389/fmicb.2021.695517. eCollection 2021. Front Microbiol. 2021. PMID: 34566906 Free PMC article.
-
Sugar transport in thermophiles: Bridging lignocellulose deconstruction and bioconversion.J Ind Microbiol Biotechnol. 2024 Jan 9;51:kuae020. doi: 10.1093/jimb/kuae020. J Ind Microbiol Biotechnol. 2024. PMID: 38866721 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

