A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6-12
- PMID: 32884066
- PMCID: PMC7471924
- DOI: 10.1038/s41598-020-71648-w
A CRISPR activation and interference toolkit for industrial Saccharomyces cerevisiae strain KE6-12
Abstract
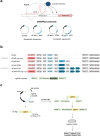
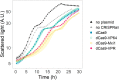
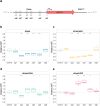
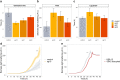
Recent advances in CRISPR/Cas9 based genome editing have considerably advanced genetic engineering of industrial yeast strains. In this study, we report the construction and characterization of a toolkit for CRISPR activation and interference (CRISPRa/i) for a polyploid industrial yeast strain. In the CRISPRa/i plasmids that are available in high and low copy variants, dCas9 is expressed alone, or as a fusion with an activation or repression domain; VP64, VPR or Mxi1. The sgRNA is introduced to the CRISPRa/i plasmids from a double stranded oligonucleotide by in vivo homology-directed repair, allowing rapid transcriptional modulation of new target genes without cloning. The CRISPRa/i toolkit was characterized by alteration of expression of fluorescent protein-encoding genes under two different promoters allowing expression alterations up to ~ 2.5-fold. Furthermore, we demonstrated the usability of the CRISPRa/i toolkit by improving the tolerance towards wheat straw hydrolysate of our industrial production strain. We anticipate that our CRISPRa/i toolkit can be widely used to assess novel targets for strain improvement and thus accelerate the design-build-test cycle for developing various industrial production strains.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Krivoruchko A, Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2015;35:7–15. - PubMed
-
- Hemansi et al. Second generation bioethanol production: The state of art. In Sustainable Approaches for Biofuels Production Technologies: From Current Status to Practical Implementation (eds. Srivastava et al.) 121–46 (Springer, Berlin, 2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

