Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters
- PMID: 32884153
- PMCID: PMC7671939
- DOI: 10.1038/s41591-020-1070-6
Ad26 vaccine protects against SARS-CoV-2 severe clinical disease in hamsters
Abstract
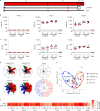
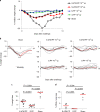
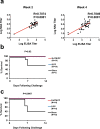
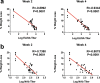
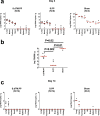
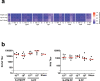
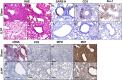
Coronavirus disease 2019 (COVID-19) in humans is often a clinically mild illness, but some individuals develop severe pneumonia, respiratory failure and death1-4. Studies of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in hamsters5-7 and nonhuman primates8-10 have generally reported mild clinical disease, and preclinical SARS-CoV-2 vaccine studies have demonstrated reduction of viral replication in the upper and lower respiratory tracts in nonhuman primates11-13. Here we show that high-dose intranasal SARS-CoV-2 infection in hamsters results in severe clinical disease, including high levels of virus replication in tissues, extensive pneumonia, weight loss and mortality in a subset of animals. A single immunization with an adenovirus serotype 26 vector-based vaccine expressing a stabilized SARS-CoV-2 spike protein elicited binding and neutralizing antibody responses and protected against SARS-CoV-2-induced weight loss, pneumonia and mortality. These data demonstrate vaccine protection against SARS-CoV-2 clinical disease. This model should prove useful for preclinical studies of SARS-CoV-2 vaccines, therapeutics and pathogenesis.
Conflict of interest statement
D.H.B., F.W., J.C., H.S. and R.Z. are co-inventors on related vaccine patents. F.W., J.C., H.S. and R.Z. are employees of Janssen Vaccines & Prevention BV and hold stock in Johnson & Johnson.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- T32 AI007387/AI/NIAID NIH HHS/United States
- P51 OD011092/OD/NIH HHS/United States
- UM1 AI126603/AI/NIAID NIH HHS/United States
- U01 CA260476/CA/NCI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- S10 OD025002/OD/NIH HHS/United States
- R01 AI146779/AI/NIAID NIH HHS/United States
- UM1 AI124377/AI/NIAID NIH HHS/United States
- U19 AI128751/AI/NIAID NIH HHS/United States
- R01 AI129797/AI/NIAID NIH HHS/United States
- K08 AI135098/AI/NIAID NIH HHS/United States
- R01 OD024917/OD/NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

