Identifying signaling genes in spatial single-cell expression data
- PMID: 32886099
- PMCID: PMC8128476
- DOI: 10.1093/bioinformatics/btaa769
Identifying signaling genes in spatial single-cell expression data
Abstract
Motivation: Recent technological advances enable the profiling of spatial single-cell expression data. Such data present a unique opportunity to study cell-cell interactions and the signaling genes that mediate them. However, most current methods for the analysis of these data focus on unsupervised descriptive modeling, making it hard to identify key signaling genes and quantitatively assess their impact.
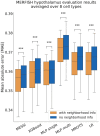
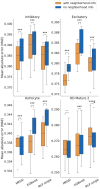
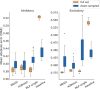
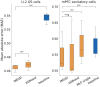
Results: We developed a Mixture of Experts for Spatial Signaling genes Identification (MESSI) method to identify active signaling genes within and between cells. The mixture of experts strategy enables MESSI to subdivide cells into subtypes. MESSI relies on multi-task learning using information from neighboring cells to improve the prediction of response genes within a cell. Applying the methods to three spatial single-cell expression datasets, we show that MESSI accurately predicts the levels of response genes, improving upon prior methods and provides useful biological insights about key signaling genes and subtypes of excitatory neuron cells.
Availability and implementation: MESSI is available at: https://github.com/doraadong/MESSI.
Supplementary information: Supplementary data are available at Bioinformatics online.
© The Author(s) 2020. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures


References
-
- Bayerl D.S. et al. (2019) More than reproduction: central gonadotropin-releasing hormone antagonism decreases maternal aggression in lactating rats. J. Neuroendocrinol., 31, e12709. - PubMed
-
- Cembrowski M.S. (2019) Single-cell transcriptomics as a framework and roadmap for understanding the brain. J.Neurosci. Methods, 326, 108353. - PubMed
-
- Chen T., Guestrin C. (2016). XGBoost: A scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. KDD ’16, p.785–794. ACM, New York, NY, USA.

